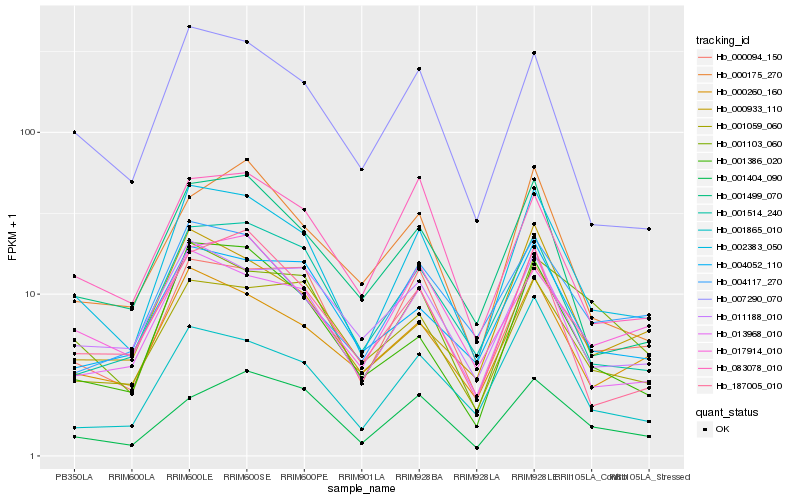
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002383_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 2 |
Hb_013968_010 |
0.0874835573 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 3 |
Hb_007290_070 |
0.1061464635 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 4 |
Hb_000260_160 |
0.1069327434 |
transcription factor |
TF Family: GNAT |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_001499_070 |
0.1079187663 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 6 |
Hb_000094_150 |
0.1123108298 |
- |
- |
- |
| 7 |
Hb_001514_240 |
0.1137440706 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 8 |
Hb_001103_060 |
0.121241669 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |
| 9 |
Hb_001386_020 |
0.1222370085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_187005_010 |
0.1226147761 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 11 |
Hb_011188_010 |
0.1244465112 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001865_010 |
0.1244795329 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 13 |
Hb_004052_110 |
0.1279920998 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001059_060 |
0.1283344618 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 15 |
Hb_083078_010 |
0.1295962902 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 16 |
Hb_004117_270 |
0.1297394114 |
- |
- |
- |
| 17 |
Hb_000933_110 |
0.1297970095 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 18 |
Hb_017914_010 |
0.1306707477 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 19 |
Hb_001404_090 |
0.1311329176 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 20 |
Hb_000175_270 |
0.1327090049 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |