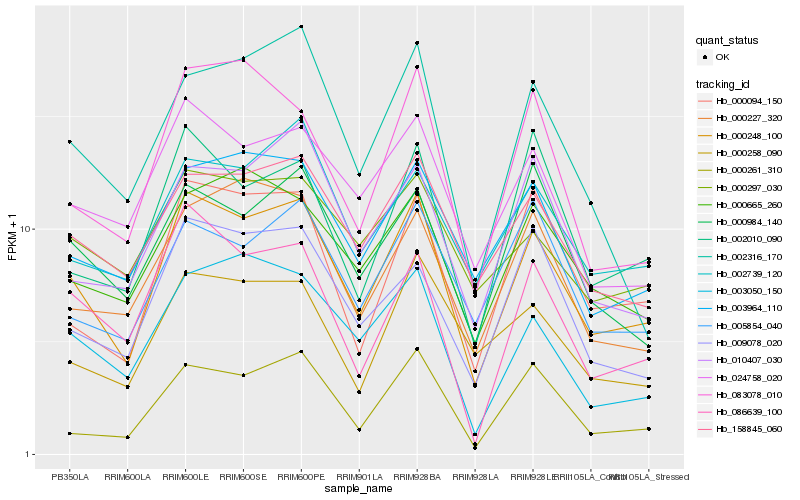
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000248_100 |
0.0 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 2 |
Hb_158845_060 |
0.1050977979 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_024758_020 |
0.1091334979 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 4 |
Hb_000094_150 |
0.1106682524 |
- |
- |
- |
| 5 |
Hb_003964_110 |
0.1117206658 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 6 |
Hb_003050_150 |
0.1173094602 |
- |
- |
endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase, putative [Ricinus communis] |
| 7 |
Hb_000261_310 |
0.1183039049 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X3 [Jatropha curcas] |
| 8 |
Hb_000665_260 |
0.1190222033 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000258_090 |
0.119920466 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 10 |
Hb_086639_100 |
0.1201021217 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105646787 [Jatropha curcas] |
| 11 |
Hb_002010_090 |
0.1205297386 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_009078_020 |
0.1207915668 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 13 |
Hb_083078_010 |
0.1214552194 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 14 |
Hb_002739_120 |
0.1223018498 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 15 |
Hb_000227_320 |
0.1233154179 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_010407_030 |
0.123781311 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 17 |
Hb_000984_140 |
0.1243950411 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 18 |
Hb_005854_040 |
0.1257286105 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000297_030 |
0.1260191889 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002316_170 |
0.1279862082 |
- |
- |
purine permease, putative [Ricinus communis] |