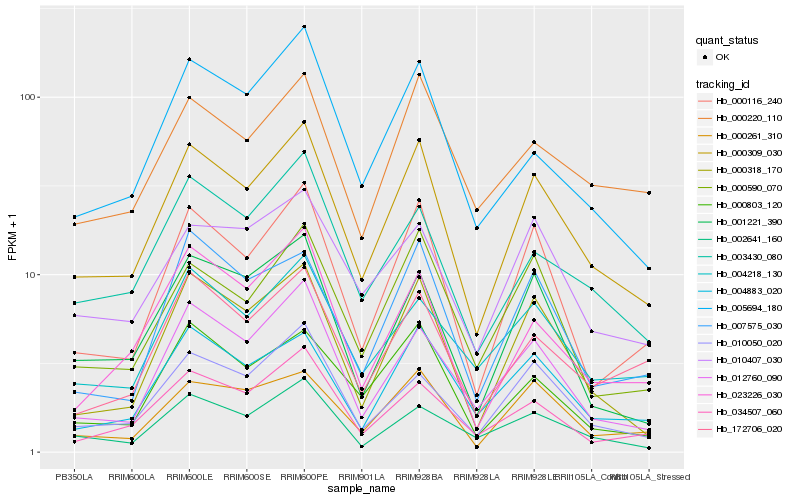
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000309_030 |
0.0 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000116_240 |
0.091494582 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 3 |
Hb_000261_310 |
0.0948939203 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X3 [Jatropha curcas] |
| 4 |
Hb_004883_020 |
0.0980974982 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 5 |
Hb_000318_170 |
0.1005803153 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 6 |
Hb_010050_020 |
0.1020409897 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 7 |
Hb_000590_070 |
0.1046582771 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 8 |
Hb_003430_080 |
0.1066309469 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_010407_030 |
0.1093939001 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 10 |
Hb_012760_090 |
0.1102602159 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 11 |
Hb_004218_130 |
0.1103406362 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 12 |
Hb_002641_160 |
0.1118006209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001221_390 |
0.1124162065 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 14 |
Hb_000803_120 |
0.1131004103 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_172706_020 |
0.1137207539 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 16 |
Hb_000220_110 |
0.1142887172 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 17 |
Hb_034507_060 |
0.1148579409 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic-like [Citrus sinensis] |
| 18 |
Hb_007575_030 |
0.116231266 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 19 |
Hb_005694_180 |
0.1175538321 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 20 |
Hb_023226_030 |
0.1220821189 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-14-like [Jatropha curcas] |