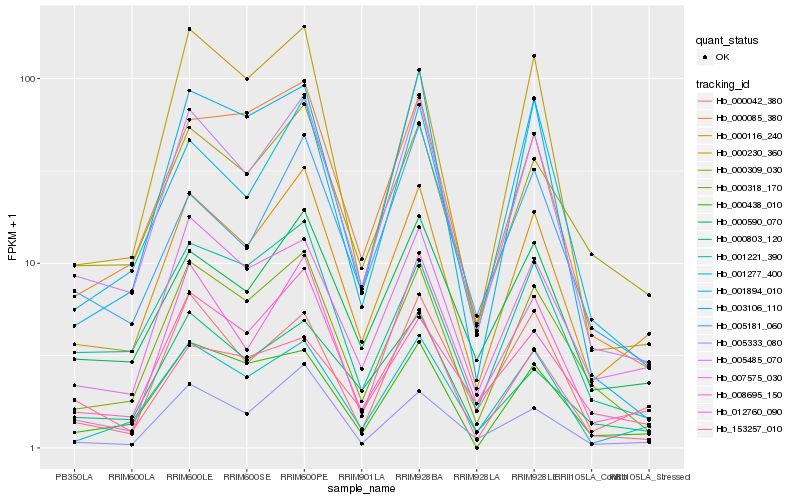
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005485_070 |
0.0 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000116_240 |
0.0794199113 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 3 |
Hb_000318_170 |
0.0892891076 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 4 |
Hb_008695_150 |
0.1007535777 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000590_070 |
0.1080231715 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 6 |
Hb_001277_400 |
0.1086903611 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 4B isoform X3 [Jatropha curcas] |
| 7 |
Hb_003106_110 |
0.1124064131 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000085_380 |
0.1166533432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007575_030 |
0.1209325651 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 10 |
Hb_001894_010 |
0.1215216737 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 11 |
Hb_005333_080 |
0.122266685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000309_030 |
0.1230300356 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_153257_010 |
0.1240290427 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 14 |
Hb_001221_390 |
0.1242705941 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 15 |
Hb_000438_010 |
0.1284169009 |
- |
- |
- |
| 16 |
Hb_000803_120 |
0.1314967404 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000042_380 |
0.1319295569 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |
| 18 |
Hb_000230_360 |
0.1322944795 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 19 |
Hb_005181_060 |
0.1328725151 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 20 |
Hb_012760_090 |
0.1337630095 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |