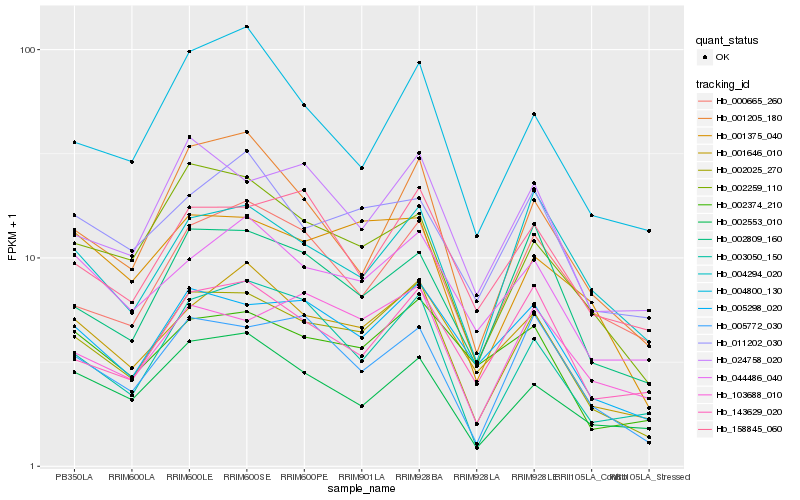
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002025_270 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g15970 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005298_020 |
0.1036056851 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 2 [Jatropha curcas] |
| 3 |
Hb_001646_010 |
0.1063784959 |
- |
- |
hypothetical protein JCGZ_16333 [Jatropha curcas] |
| 4 |
Hb_005772_030 |
0.1088503843 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 5 |
Hb_024758_020 |
0.1120112705 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 6 |
Hb_002809_160 |
0.1136794319 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 7 |
Hb_003050_150 |
0.1161836522 |
- |
- |
endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase, putative [Ricinus communis] |
| 8 |
Hb_001375_040 |
0.1178950703 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 9 |
Hb_002259_110 |
0.1180399443 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 10 |
Hb_103688_010 |
0.1211685464 |
- |
- |
PREDICTED: sucrose-phosphatase 2-like [Jatropha curcas] |
| 11 |
Hb_001205_180 |
0.1213760281 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 12 |
Hb_044486_040 |
0.1232027126 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |
| 13 |
Hb_143629_020 |
0.1241743842 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 14 |
Hb_004294_020 |
0.1255050178 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002374_210 |
0.1258693772 |
- |
- |
PREDICTED: uncharacterized protein LOC105638976 [Jatropha curcas] |
| 16 |
Hb_002553_010 |
0.1293786909 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 2-like [Populus euphratica] |
| 17 |
Hb_158845_060 |
0.1311080822 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000665_260 |
0.1311090837 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 19 |
Hb_011202_030 |
0.1314971223 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 20 |
Hb_004800_130 |
0.1331158705 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |