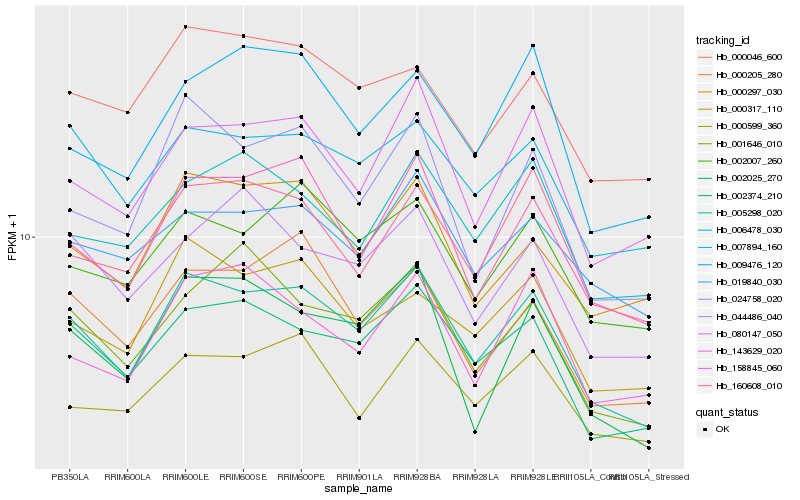
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005298_020 |
0.0 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 2 [Jatropha curcas] |
| 2 |
Hb_002374_210 |
0.0770553018 |
- |
- |
PREDICTED: uncharacterized protein LOC105638976 [Jatropha curcas] |
| 3 |
Hb_158845_060 |
0.0830912787 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_024758_020 |
0.0865151923 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 5 |
Hb_080147_050 |
0.091698501 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 6 |
Hb_002007_260 |
0.0967436144 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 7 |
Hb_007894_160 |
0.0991082974 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 8 |
Hb_000317_110 |
0.09960189 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B1, mitochondrial isoform X2 [Jatropha curcas] |
| 9 |
Hb_160608_010 |
0.1000796014 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 10 |
Hb_000599_360 |
0.1018390234 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 11 |
Hb_000297_030 |
0.1024100611 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000205_280 |
0.1028599459 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_002025_270 |
0.1036056851 |
- |
- |
PREDICTED: uncharacterized protein At4g15970 isoform X1 [Jatropha curcas] |
| 14 |
Hb_019840_030 |
0.1072487679 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001646_010 |
0.1077835595 |
- |
- |
hypothetical protein JCGZ_16333 [Jatropha curcas] |
| 16 |
Hb_006478_030 |
0.1090932896 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 17 |
Hb_000046_600 |
0.1091931561 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 18 |
Hb_009476_120 |
0.109232872 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 19 |
Hb_044486_040 |
0.1111510628 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |
| 20 |
Hb_143629_020 |
0.1113715703 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |