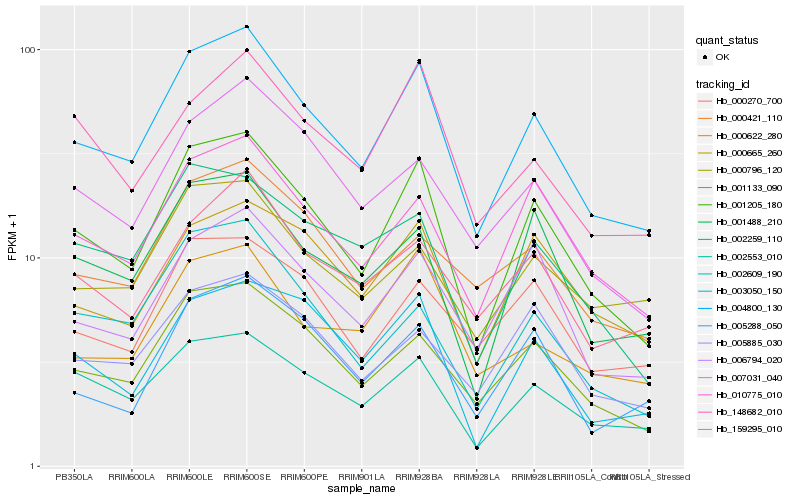
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004800_130 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 2 |
Hb_001205_180 |
0.048835388 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 3 |
Hb_001133_090 |
0.0807864237 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 4 |
Hb_002553_010 |
0.088168594 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 2-like [Populus euphratica] |
| 5 |
Hb_010775_010 |
0.0920370824 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006794_020 |
0.0974109234 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 7 |
Hb_159295_010 |
0.1003993766 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 8 |
Hb_001488_210 |
0.1028351308 |
- |
- |
PREDICTED: protein RIK [Jatropha curcas] |
| 9 |
Hb_000622_280 |
0.103222895 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007031_040 |
0.1036573716 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 11 |
Hb_000270_700 |
0.1056419313 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 12 |
Hb_000421_110 |
0.1067335359 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_148682_010 |
0.1081584546 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 14 |
Hb_000665_260 |
0.1086266265 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000796_120 |
0.1089162896 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 16 |
Hb_005288_050 |
0.1104911467 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 17 |
Hb_005885_030 |
0.1112780125 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 18 |
Hb_002259_110 |
0.1116853731 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 19 |
Hb_003050_150 |
0.1118573536 |
- |
- |
endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase, putative [Ricinus communis] |
| 20 |
Hb_002609_190 |
0.1137499689 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |