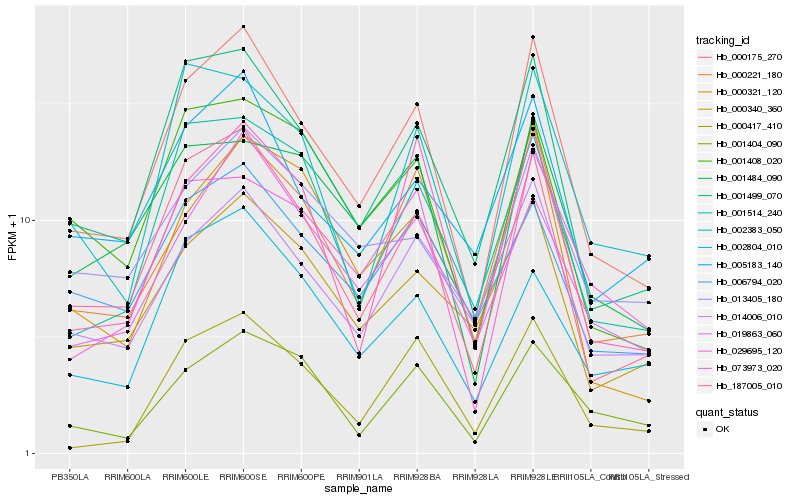
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_270 |
0.0 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 2 |
Hb_187005_010 |
0.0727130842 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 3 |
Hb_001499_070 |
0.0842368882 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 4 |
Hb_000221_180 |
0.1026251994 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 5 |
Hb_014006_010 |
0.1151950472 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 6 |
Hb_019863_060 |
0.1195158739 |
- |
- |
PREDICTED: target of Myb protein 1-like [Populus euphratica] |
| 7 |
Hb_000340_360 |
0.1200537723 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 8 |
Hb_005183_140 |
0.1221145172 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 9 |
Hb_001514_240 |
0.1233287178 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 10 |
Hb_001408_020 |
0.1253518099 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 11 |
Hb_073973_020 |
0.1256322367 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 12 |
Hb_000321_120 |
0.1273228385 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 13 |
Hb_029695_120 |
0.1281812682 |
- |
- |
hypothetical protein POPTR_0002s05320g [Populus trichocarpa] |
| 14 |
Hb_006794_020 |
0.1305882179 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 15 |
Hb_000417_410 |
0.1308016845 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001484_090 |
0.1309487606 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_001404_090 |
0.1313040533 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 18 |
Hb_002383_050 |
0.1327090049 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 19 |
Hb_013405_180 |
0.1334863296 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 20 |
Hb_002804_010 |
0.1335071242 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |