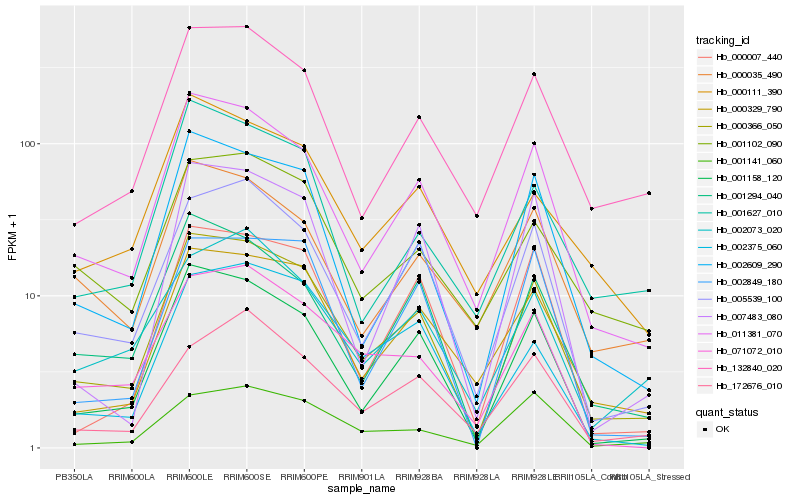
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000366_050 |
0.0 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_011381_070 |
0.0767692451 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 3 |
Hb_001158_120 |
0.080674697 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002609_290 |
0.0930373113 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 5 |
Hb_005539_100 |
0.1029585662 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 6 |
Hb_000329_790 |
0.111877621 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 7 |
Hb_001294_040 |
0.1140284946 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 8 |
Hb_132840_020 |
0.1173335123 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 9 |
Hb_071072_010 |
0.1240981182 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 10 |
Hb_000007_440 |
0.1292027683 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 11 |
Hb_002849_180 |
0.132585976 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 12 |
Hb_007483_080 |
0.1345175851 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 13 |
Hb_000111_390 |
0.1362397238 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002375_060 |
0.1366855674 |
- |
- |
PREDICTED: uncharacterized protein LOC105628271 [Jatropha curcas] |
| 15 |
Hb_002073_020 |
0.1369887959 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 16 |
Hb_000035_490 |
0.1372164646 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 17 |
Hb_001102_090 |
0.1376589568 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 18 |
Hb_001141_060 |
0.1380820997 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001627_010 |
0.1385993833 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 20 |
Hb_172676_010 |
0.1422157798 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |