| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
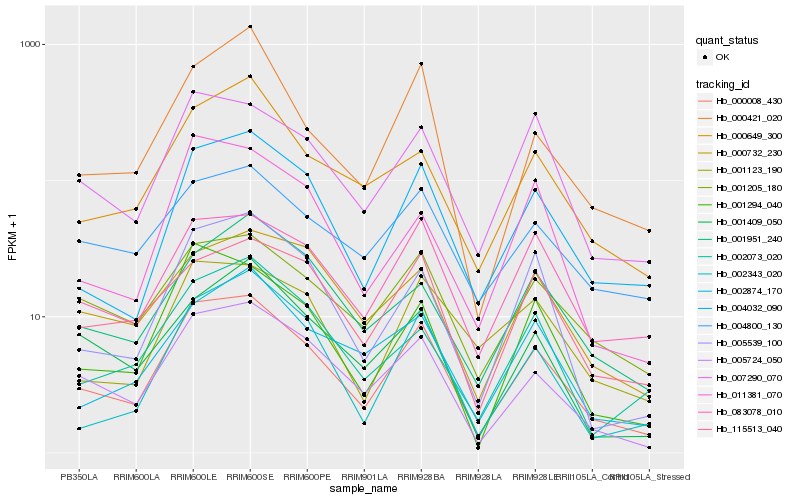
Hb_000008_430 |
0.0 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005724_050 |
0.0979597144 |
- |
- |
- |
| 3 |
Hb_004032_090 |
0.1046817556 |
- |
- |
PREDICTED: protein AIG1 [Jatropha curcas] |
| 4 |
Hb_001294_040 |
0.1195306746 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 5 |
Hb_007290_070 |
0.1223573359 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 6 |
Hb_005539_100 |
0.1226288399 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 7 |
Hb_001205_180 |
0.1240393725 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 8 |
Hb_083078_010 |
0.1241956086 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 9 |
Hb_000421_020 |
0.132677434 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 10 |
Hb_115513_040 |
0.1330623776 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000649_300 |
0.1339705553 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 12 |
Hb_002874_170 |
0.1398672328 |
- |
- |
PREDICTED: L-arabinokinase-like isoform X3 [Jatropha curcas] |
| 13 |
Hb_001123_190 |
0.1402862096 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 14 |
Hb_002343_020 |
0.1405148214 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 15 |
Hb_004800_130 |
0.1431345919 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 16 |
Hb_011381_070 |
0.1439729662 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 17 |
Hb_002073_020 |
0.1447057265 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 18 |
Hb_000732_230 |
0.1457071102 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001951_240 |
0.1457120671 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 20 |
Hb_001409_050 |
0.1464381433 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |