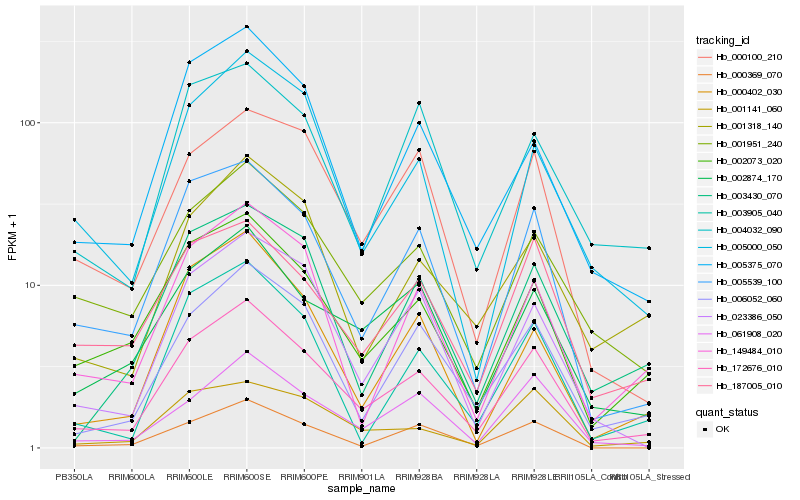
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172676_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 2 |
Hb_005539_100 |
0.0915976052 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 3 |
Hb_006052_060 |
0.1062599625 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 4 |
Hb_002073_020 |
0.1080290629 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 5 |
Hb_002874_170 |
0.1165097422 |
- |
- |
PREDICTED: L-arabinokinase-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_061908_020 |
0.1178657361 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 7 |
Hb_001951_240 |
0.1217317632 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 8 |
Hb_023386_050 |
0.127764348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001141_060 |
0.1315529338 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000100_210 |
0.131857559 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 11 |
Hb_187005_010 |
0.1319113064 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 12 |
Hb_005375_070 |
0.1320451347 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 13 |
Hb_149484_010 |
0.132706539 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor-like [Eucalyptus grandis] |
| 14 |
Hb_000369_070 |
0.1335751172 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 15 |
Hb_003905_040 |
0.1341549126 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 16 |
Hb_005000_050 |
0.1346954042 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 17 |
Hb_001318_140 |
0.1356898325 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 18 |
Hb_003430_070 |
0.1361322952 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 19 |
Hb_000402_030 |
0.1365964401 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 20 |
Hb_004032_090 |
0.1414899258 |
- |
- |
PREDICTED: protein AIG1 [Jatropha curcas] |