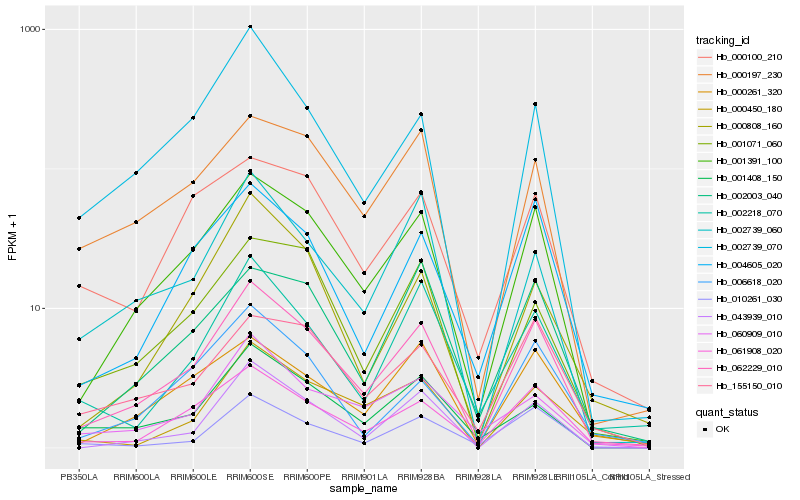
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_062229_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 2 |
Hb_001391_100 |
0.0632406433 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 3 |
Hb_061908_020 |
0.1036752718 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 4 |
Hb_004605_020 |
0.1247942265 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 5 |
Hb_043939_010 |
0.1311818271 |
- |
- |
hypothetical protein B456_N011100 [Gossypium raimondii] |
| 6 |
Hb_000197_230 |
0.1397342178 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 7 |
Hb_010261_030 |
0.1402927984 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 8 |
Hb_000261_320 |
0.1410930116 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_002739_070 |
0.142593859 |
- |
- |
hypothetical protein B456_002G169900 [Gossypium raimondii] |
| 10 |
Hb_002218_070 |
0.1458581779 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 11 |
Hb_002003_040 |
0.1471564155 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002739_060 |
0.1492764574 |
- |
- |
- |
| 13 |
Hb_155150_010 |
0.1493223175 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_001408_150 |
0.1505184407 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001071_060 |
0.1538756193 |
- |
- |
Trehalose phosphatase/synthase 5 isoform 1 [Theobroma cacao] |
| 16 |
Hb_060909_010 |
0.1539277772 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 17 |
Hb_000450_180 |
0.1541088349 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 18 |
Hb_000100_210 |
0.1541114952 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 19 |
Hb_006618_020 |
0.155044626 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 20 |
Hb_000808_160 |
0.1560921805 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |