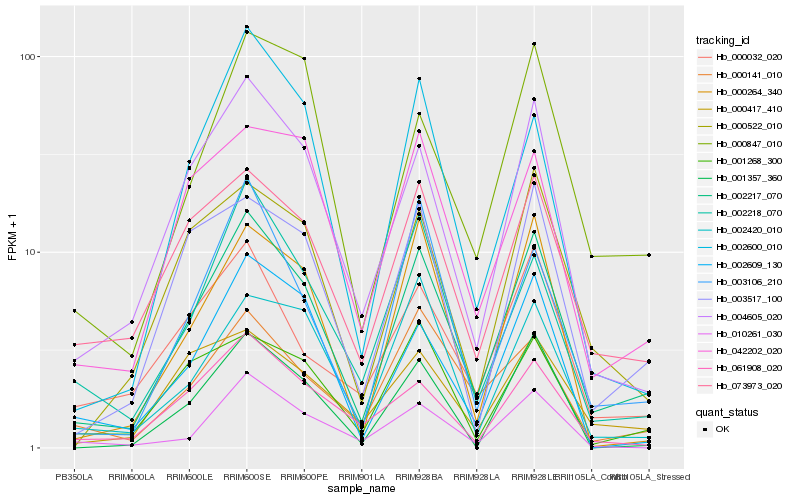
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643596 [Jatropha curcas] |
| 2 |
Hb_004605_020 |
0.1166395375 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 3 |
Hb_073973_020 |
0.1372169699 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 4 |
Hb_002218_070 |
0.1435247454 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 5 |
Hb_003517_100 |
0.144825587 |
transcription factor |
TF Family: GRAS |
- |
| 6 |
Hb_000522_010 |
0.1457541003 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 7 |
Hb_000847_010 |
0.1469541643 |
- |
- |
PREDICTED: uncharacterized protein LOC102608064 [Citrus sinensis] |
| 8 |
Hb_061908_020 |
0.1482096025 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 9 |
Hb_000264_340 |
0.1505272169 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_042202_020 |
0.1513459463 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 11 |
Hb_001268_300 |
0.1528964851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003106_210 |
0.1529485894 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |
| 13 |
Hb_002609_130 |
0.1532274763 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 14 |
Hb_010261_030 |
0.1540028754 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 15 |
Hb_000032_020 |
0.1540805214 |
- |
- |
Centromeric protein E, putative [Ricinus communis] |
| 16 |
Hb_001357_360 |
0.1549656769 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_002600_010 |
0.1550136106 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000417_410 |
0.156007714 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000141_010 |
0.1570675233 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 20 |
Hb_002420_010 |
0.1589051472 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |