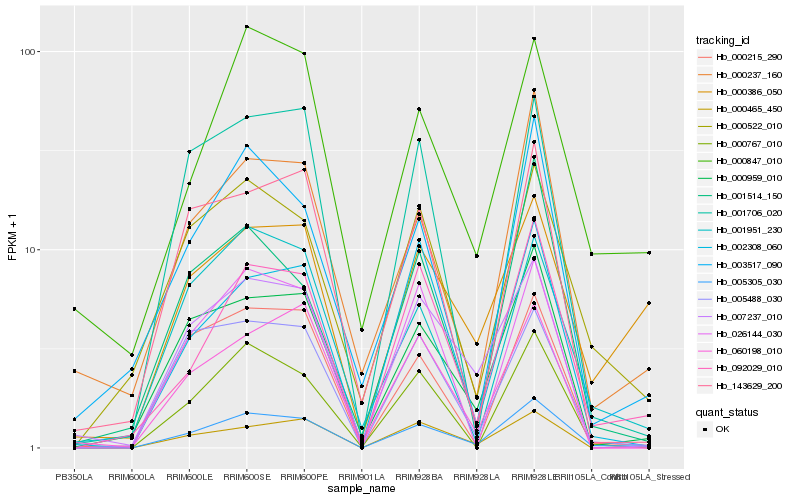
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005305_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_25922 [Jatropha curcas] |
| 2 |
Hb_092029_010 |
0.1201998275 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 3 |
Hb_000959_010 |
0.1229384251 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000465_450 |
0.1326006161 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 5 |
Hb_000767_010 |
0.1383996106 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 6 |
Hb_000522_010 |
0.149500587 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 7 |
Hb_007237_010 |
0.1510290536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000237_160 |
0.1528470567 |
- |
- |
BEL1-like homeodomain protein 1 [Morus notabilis] |
| 9 |
Hb_000215_290 |
0.1534459939 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_005488_030 |
0.1573021965 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 11 |
Hb_002308_060 |
0.160986213 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 12 |
Hb_003517_090 |
0.1615463161 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001706_020 |
0.1616956831 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 14 |
Hb_000847_010 |
0.1618589094 |
- |
- |
PREDICTED: uncharacterized protein LOC102608064 [Citrus sinensis] |
| 15 |
Hb_001951_230 |
0.1638595868 |
- |
- |
PREDICTED: phytochrome E isoform X2 [Jatropha curcas] |
| 16 |
Hb_026144_030 |
0.1654497688 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 17 |
Hb_000386_050 |
0.1655784909 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_060198_010 |
0.169313853 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 19 |
Hb_143629_200 |
0.1699546699 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 20 |
Hb_001514_150 |
0.171460537 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |