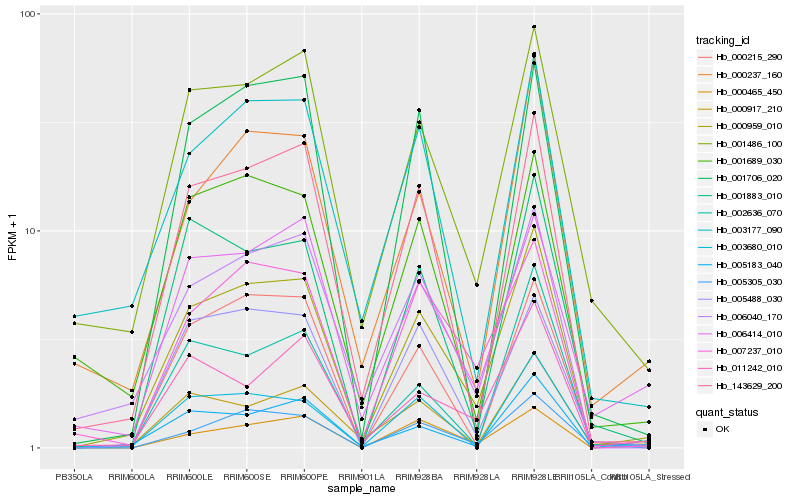
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000959_010 |
0.0 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_143629_200 |
0.1069948538 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 3 |
Hb_001486_100 |
0.1207259033 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 4 |
Hb_006040_170 |
0.1212772438 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |
| 5 |
Hb_005305_030 |
0.1229384251 |
- |
- |
hypothetical protein JCGZ_25922 [Jatropha curcas] |
| 6 |
Hb_007237_010 |
0.1261750367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000237_160 |
0.1305568372 |
- |
- |
BEL1-like homeodomain protein 1 [Morus notabilis] |
| 8 |
Hb_000215_290 |
0.1311114448 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005183_040 |
0.1331817122 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 10 |
Hb_005488_030 |
0.1338894265 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 11 |
Hb_003177_090 |
0.1353427888 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 12 |
Hb_000917_210 |
0.1368893838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001883_010 |
0.1375277532 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 14 |
Hb_000465_450 |
0.1383805072 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 15 |
Hb_011242_010 |
0.1402201025 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 16 |
Hb_003680_010 |
0.1410663609 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_006414_010 |
0.1438993875 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 18 |
Hb_002636_070 |
0.1453680517 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 19 |
Hb_001706_020 |
0.1462757021 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 20 |
Hb_001689_030 |
0.1462931321 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |