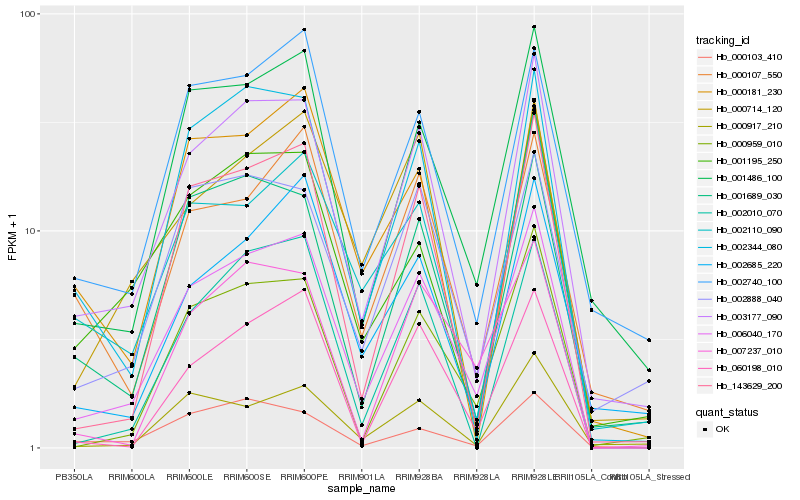
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006040_170 |
0.0 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |
| 2 |
Hb_003177_090 |
0.0890305359 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 3 |
Hb_143629_200 |
0.113944712 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 4 |
Hb_001486_100 |
0.1161623598 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 5 |
Hb_000959_010 |
0.1212772438 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_060198_010 |
0.1269801429 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 7 |
Hb_002344_080 |
0.1291788046 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 8 |
Hb_002010_070 |
0.1347675371 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |
| 9 |
Hb_007237_010 |
0.1348918933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001689_030 |
0.1353344792 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 11 |
Hb_002888_040 |
0.1386078211 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 12 |
Hb_002740_100 |
0.1400336706 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 13 |
Hb_002110_090 |
0.1424452001 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 14 |
Hb_002685_220 |
0.1487153787 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000714_120 |
0.1493238168 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 16 |
Hb_000107_550 |
0.1498699094 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 17 |
Hb_001195_250 |
0.1502097007 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000917_210 |
0.1522152561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000103_410 |
0.1530143858 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 20 |
Hb_000181_230 |
0.1553282567 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |