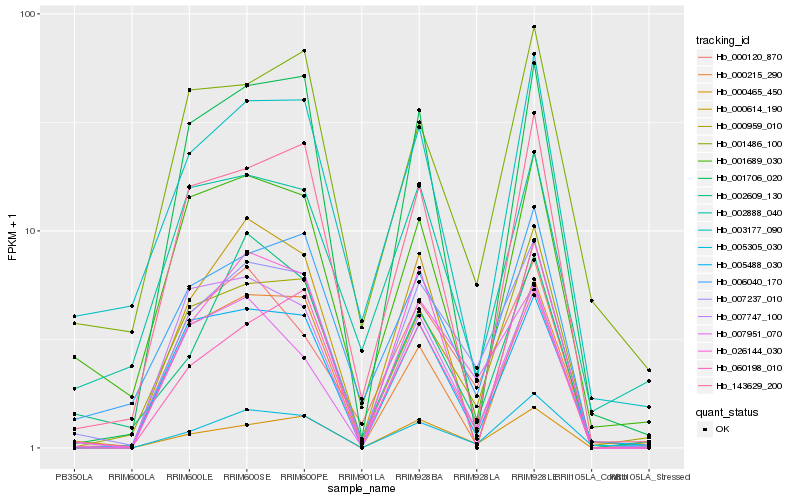
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007237_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000959_010 |
0.1261750367 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_060198_010 |
0.1311393444 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 4 |
Hb_006040_170 |
0.1348918933 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |
| 5 |
Hb_000120_870 |
0.1353558494 |
rubber biosynthesis |
Gene Name: CPT6 |
- |
| 6 |
Hb_000465_450 |
0.1356151247 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 7 |
Hb_007951_070 |
0.1425484241 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 8 |
Hb_005488_030 |
0.1504150759 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 9 |
Hb_005305_030 |
0.1510290536 |
- |
- |
hypothetical protein JCGZ_25922 [Jatropha curcas] |
| 10 |
Hb_143629_200 |
0.1607562725 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 11 |
Hb_007747_100 |
0.1649043903 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 12 |
Hb_001706_020 |
0.1684208293 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 13 |
Hb_003177_090 |
0.1693025113 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 14 |
Hb_002609_130 |
0.1703613841 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 15 |
Hb_001486_100 |
0.1703712724 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 16 |
Hb_001689_030 |
0.1721125418 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 17 |
Hb_026144_030 |
0.1739394729 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 18 |
Hb_000215_290 |
0.1746960281 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002888_040 |
0.1757845811 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 20 |
Hb_000614_190 |
0.1758053373 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |