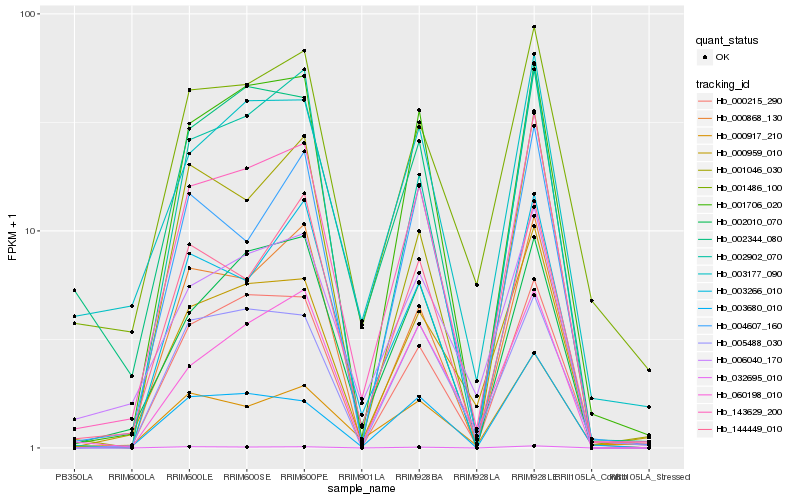
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143629_200 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 2 |
Hb_001706_020 |
0.0862125034 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 3 |
Hb_000215_290 |
0.0953149519 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002010_070 |
0.0992688263 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |
| 5 |
Hb_144449_010 |
0.1055958161 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 6 |
Hb_000868_130 |
0.1064519741 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 7 |
Hb_000959_010 |
0.1069948538 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_006040_170 |
0.113944712 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |
| 9 |
Hb_002344_080 |
0.1146587031 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 10 |
Hb_060198_010 |
0.1148054735 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 11 |
Hb_003177_090 |
0.1151532893 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 12 |
Hb_002902_070 |
0.1163793666 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 13 |
Hb_004607_160 |
0.1176049855 |
- |
- |
PREDICTED: uncharacterized protein LOC105134931 [Populus euphratica] |
| 14 |
Hb_005488_030 |
0.1180233402 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 15 |
Hb_003266_010 |
0.1186267242 |
- |
- |
hypothetical protein JCGZ_21945 [Jatropha curcas] |
| 16 |
Hb_032695_010 |
0.1186356239 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Vitis vinifera] |
| 17 |
Hb_001046_030 |
0.1235162227 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_003680_010 |
0.1235867171 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_001486_100 |
0.124437864 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 20 |
Hb_000917_210 |
0.124494629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |