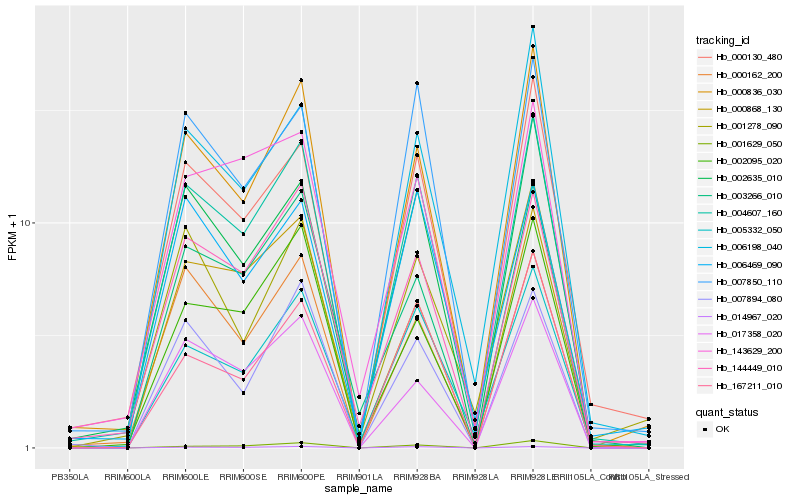
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004607_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105134931 [Populus euphratica] |
| 2 |
Hb_000836_030 |
0.0700774326 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 3 |
Hb_000130_480 |
0.0799941186 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 4 |
Hb_002635_010 |
0.0820229686 |
- |
- |
homogentisate phytyl transferase [Hevea brasiliensis] |
| 5 |
Hb_144449_010 |
0.0882601111 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 6 |
Hb_005332_050 |
0.0894301 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 7 |
Hb_007850_110 |
0.0904916035 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 8 |
Hb_006198_040 |
0.105755774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_014967_020 |
0.1074770028 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_000162_200 |
0.1075136482 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 11 |
Hb_007894_080 |
0.1126608886 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_167211_010 |
0.1141436409 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002095_020 |
0.1147761836 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003266_010 |
0.1165106276 |
- |
- |
hypothetical protein JCGZ_21945 [Jatropha curcas] |
| 15 |
Hb_143629_200 |
0.1176049855 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 16 |
Hb_001629_050 |
0.1185149901 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 17 |
Hb_017358_020 |
0.1196040324 |
- |
- |
hypothetical protein POPTR_0014s16490g [Populus trichocarpa] |
| 18 |
Hb_000868_130 |
0.120429188 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 19 |
Hb_006469_090 |
0.1217100175 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 20 |
Hb_001278_090 |
0.1225685211 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |