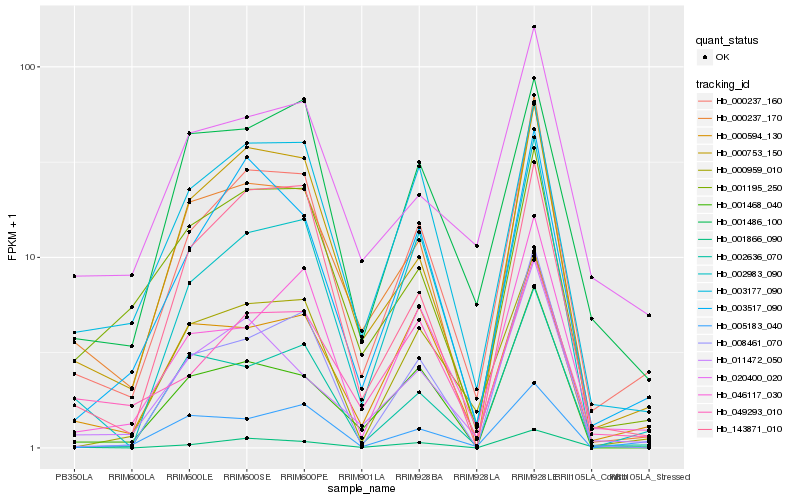
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000237_160 |
0.0 |
- |
- |
BEL1-like homeodomain protein 1 [Morus notabilis] |
| 2 |
Hb_000753_150 |
0.1043736744 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 3 |
Hb_003517_090 |
0.1106099347 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_002983_090 |
0.1161110678 |
- |
- |
Altered inheritance of mitochondria protein 32 [Glycine soja] |
| 5 |
Hb_003177_090 |
0.1214647237 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 6 |
Hb_000237_170 |
0.1246180371 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 6 [Jatropha curcas] |
| 7 |
Hb_000959_010 |
0.1305568372 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_046117_030 |
0.1316717119 |
- |
- |
sialyltransferase, putative [Ricinus communis] |
| 9 |
Hb_001866_090 |
0.1354223381 |
- |
- |
hypothetical protein VITISV_023491 [Vitis vinifera] |
| 10 |
Hb_000594_130 |
0.1368952794 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_143871_010 |
0.1417292876 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 12 |
Hb_011472_050 |
0.1424667607 |
- |
- |
- |
| 13 |
Hb_008461_070 |
0.1450615303 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005183_040 |
0.1462895292 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 15 |
Hb_002636_070 |
0.1471596537 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 16 |
Hb_049293_010 |
0.1475066186 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_001195_250 |
0.1509092636 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_001486_100 |
0.1511686389 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 19 |
Hb_020400_020 |
0.1521307589 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 20 |
Hb_001468_040 |
0.1522815251 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |