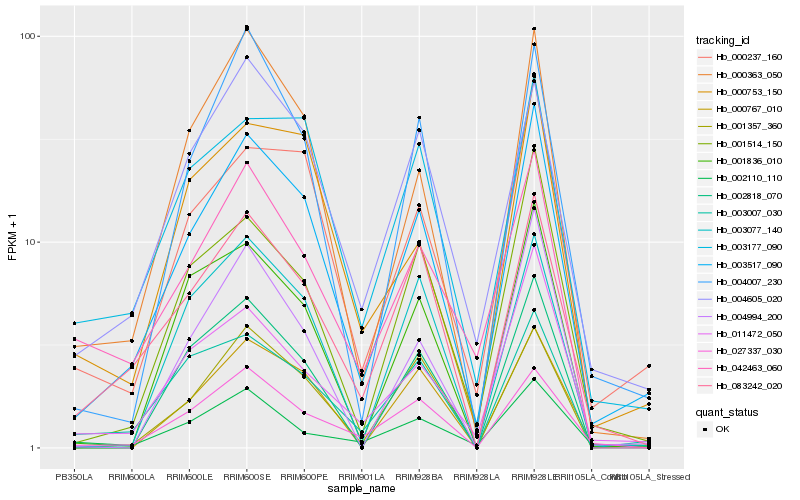
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003517_090 |
0.0 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000237_160 |
0.1106099347 |
- |
- |
BEL1-like homeodomain protein 1 [Morus notabilis] |
| 3 |
Hb_000363_050 |
0.1184730533 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_083242_020 |
0.1272955799 |
- |
- |
hypothetical protein JCGZ_20097 [Jatropha curcas] |
| 5 |
Hb_001514_150 |
0.1305291323 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 6 |
Hb_011472_050 |
0.131427347 |
- |
- |
- |
| 7 |
Hb_004605_020 |
0.1353204696 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 8 |
Hb_004007_230 |
0.1360552936 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003177_090 |
0.1373795723 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 10 |
Hb_001357_360 |
0.1378511877 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000753_150 |
0.138179368 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 12 |
Hb_027337_030 |
0.1415150033 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |
| 13 |
Hb_003007_030 |
0.1427730291 |
- |
- |
- |
| 14 |
Hb_001836_010 |
0.1449776037 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 15 |
Hb_000767_010 |
0.1454086174 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 16 |
Hb_004994_200 |
0.1459056933 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 17 |
Hb_002110_110 |
0.1516728704 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 18 |
Hb_002818_070 |
0.1532062213 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 19 |
Hb_003077_140 |
0.153982559 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 20 |
Hb_042463_060 |
0.155445013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |