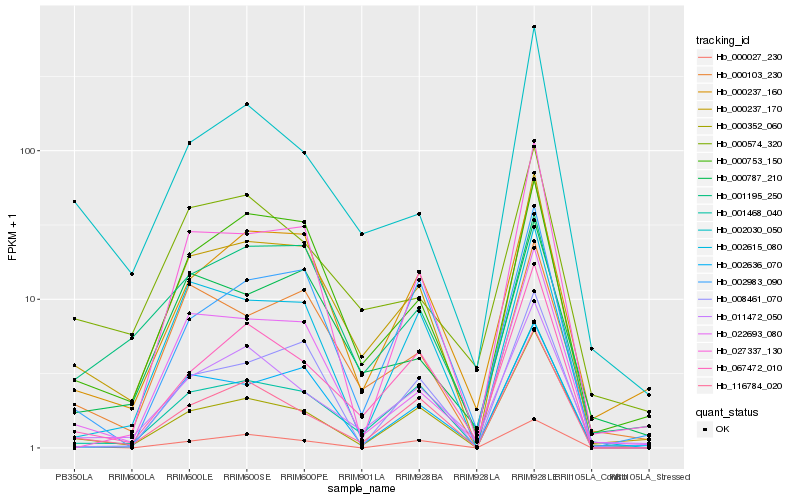
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000237_170 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 6 [Jatropha curcas] |
| 2 |
Hb_001468_040 |
0.1030126693 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 3 |
Hb_000753_150 |
0.115635456 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 4 |
Hb_000103_230 |
0.1206137455 |
- |
- |
PREDICTED: chlorophyllase-2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000237_160 |
0.1246180371 |
- |
- |
BEL1-like homeodomain protein 1 [Morus notabilis] |
| 6 |
Hb_022693_080 |
0.1249702169 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 7 |
Hb_011472_050 |
0.1353827588 |
- |
- |
- |
| 8 |
Hb_067472_010 |
0.1361466775 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 9 |
Hb_116784_020 |
0.137074145 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_002615_080 |
0.1373244424 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 11 |
Hb_002636_070 |
0.1394661078 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 12 |
Hb_000787_210 |
0.1400957203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000352_060 |
0.1437901908 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 14 |
Hb_002983_090 |
0.1487921304 |
- |
- |
Altered inheritance of mitochondria protein 32 [Glycine soja] |
| 15 |
Hb_008461_070 |
0.1491754078 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000027_230 |
0.1503584628 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 17 |
Hb_027337_130 |
0.152177866 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 18 |
Hb_000574_320 |
0.1529552343 |
- |
- |
hypothetical protein CICLE_v10024937mg [Citrus clementina] |
| 19 |
Hb_002030_050 |
0.1536606831 |
- |
- |
classical arabinogalactan protein 7 precursor [Jatropha curcas] |
| 20 |
Hb_001195_250 |
0.1577599266 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |