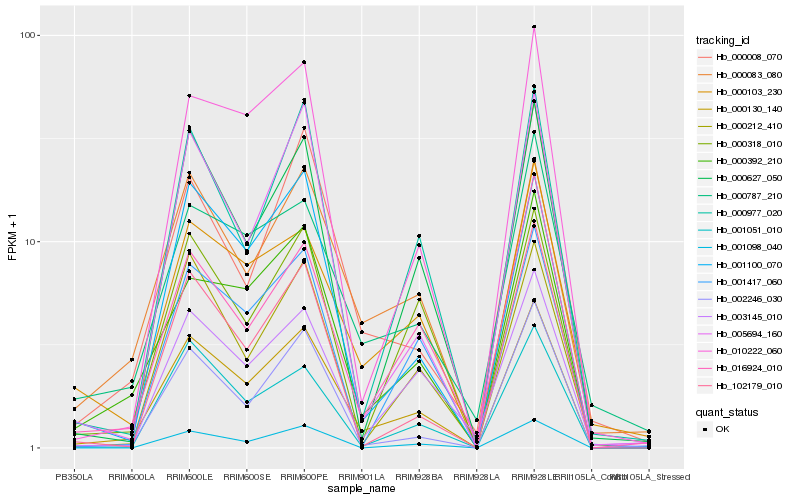
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_140 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639203 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000392_210 |
0.1048004816 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0004s04600g [Populus trichocarpa] |
| 3 |
Hb_000083_080 |
0.1086022737 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 4 |
Hb_016924_010 |
0.1191245622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000103_230 |
0.1195849 |
- |
- |
PREDICTED: chlorophyllase-2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001100_070 |
0.1218321385 |
- |
- |
PREDICTED: ferredoxin--nitrite reductase, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000627_050 |
0.1235949972 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 8 |
Hb_010222_060 |
0.1263973789 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000977_020 |
0.1282909134 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 10 |
Hb_000787_210 |
0.1290622426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003145_010 |
0.1295492059 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 12 |
Hb_001051_010 |
0.1300591358 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 13 |
Hb_000212_410 |
0.1301041871 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 14 |
Hb_001417_060 |
0.1316986241 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 15 |
Hb_002246_030 |
0.1320566718 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 16 |
Hb_001098_040 |
0.132160415 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 17 |
Hb_000318_010 |
0.1336204517 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 18 |
Hb_000008_070 |
0.1337501282 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_005694_160 |
0.1339408396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_102179_010 |
0.1343103301 |
- |
- |
protein binding protein, putative [Ricinus communis] |