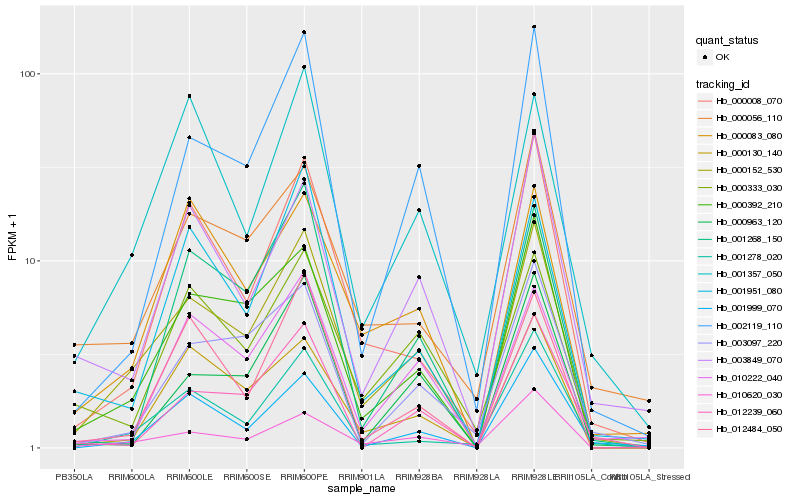
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_530 |
0.0 |
- |
- |
hypothetical protein SELMODRAFT_231485 [Selaginella moellendorffii] |
| 2 |
Hb_000392_210 |
0.1146333267 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0004s04600g [Populus trichocarpa] |
| 3 |
Hb_002119_110 |
0.1329766078 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 4 |
Hb_000008_070 |
0.1475269601 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_000130_140 |
0.1502610724 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639203 isoform X1 [Jatropha curcas] |
| 6 |
Hb_010620_030 |
0.1506904122 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 7 |
Hb_000056_110 |
0.1509862372 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 8 |
Hb_001999_070 |
0.1537348966 |
- |
- |
PREDICTED: ammonium transporter 1 member 3 [Jatropha curcas] |
| 9 |
Hb_001278_020 |
0.1549243096 |
- |
- |
sucrose synthase 6 [Hevea brasiliensis] |
| 10 |
Hb_000083_080 |
0.1575180438 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 11 |
Hb_010222_040 |
0.1586011046 |
- |
- |
PREDICTED: kinesin-2 [Jatropha curcas] |
| 12 |
Hb_003849_070 |
0.1586918293 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 13 |
Hb_000333_030 |
0.1600860817 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003097_220 |
0.1601322762 |
- |
- |
hypothetical protein POPTR_0001s06150g [Populus trichocarpa] |
| 15 |
Hb_012484_050 |
0.1610773532 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 16 |
Hb_000963_120 |
0.1617293659 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001951_080 |
0.1623505013 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 18 |
Hb_001357_050 |
0.1651575846 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12-like [Jatropha curcas] |
| 19 |
Hb_012239_060 |
0.1653024116 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001268_150 |
0.1672938604 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |