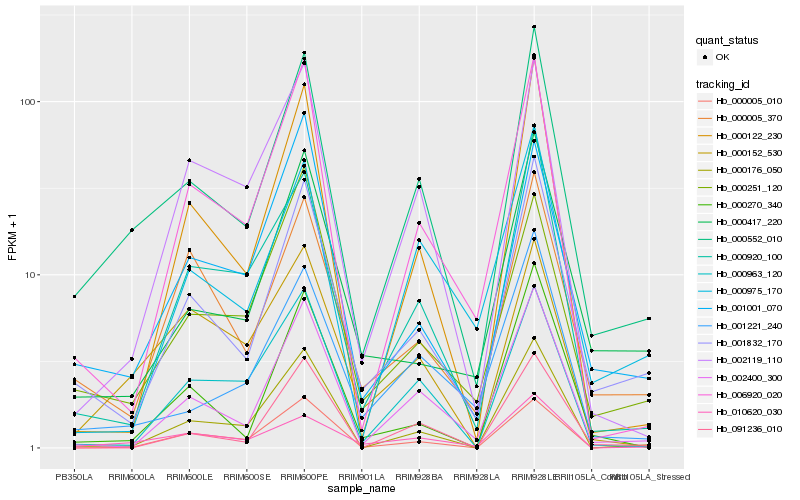
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000552_010 |
0.0 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 2 [Jatropha curcas] |
| 2 |
Hb_001832_170 |
0.1191989399 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 3 |
Hb_002400_300 |
0.1347395144 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 4 |
Hb_010620_030 |
0.1427453845 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 5 |
Hb_000920_100 |
0.1476501393 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000176_050 |
0.148337663 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 7 |
Hb_001221_240 |
0.1502537641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006920_020 |
0.1599267712 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 9 |
Hb_000122_230 |
0.1613094432 |
- |
- |
laccase, putative [Ricinus communis] |
| 10 |
Hb_002119_110 |
0.1635693674 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 11 |
Hb_000005_370 |
0.1651736641 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634024 [Jatropha curcas] |
| 12 |
Hb_000975_170 |
0.167977176 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000417_220 |
0.1681367529 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Cucumis melo] |
| 14 |
Hb_000963_120 |
0.16818607 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000152_530 |
0.169771313 |
- |
- |
hypothetical protein SELMODRAFT_231485 [Selaginella moellendorffii] |
| 16 |
Hb_000270_340 |
0.1727100016 |
- |
- |
hypothetical protein JCGZ_11546 [Jatropha curcas] |
| 17 |
Hb_000005_010 |
0.1736525417 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 18 |
Hb_000251_120 |
0.1745223316 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 19 |
Hb_001001_070 |
0.1762042405 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 20 |
Hb_091236_010 |
0.1767380348 |
- |
- |
hypothetical protein POPTR_0019s12290g [Populus trichocarpa] |