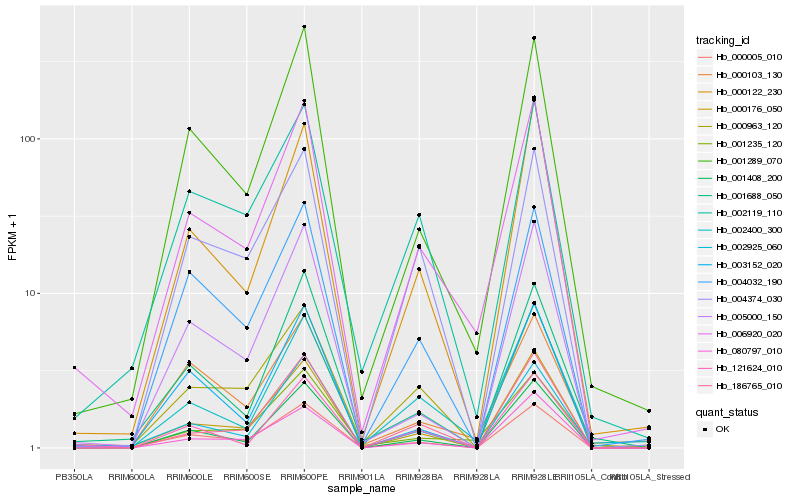
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006920_020 |
0.0 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 2 |
Hb_001289_070 |
0.0980740519 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 3 |
Hb_000176_050 |
0.0982294667 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 4 |
Hb_002400_300 |
0.1038371686 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 5 |
Hb_000122_230 |
0.1081072402 |
- |
- |
laccase, putative [Ricinus communis] |
| 6 |
Hb_002925_060 |
0.1105687851 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 7 |
Hb_001408_200 |
0.1112801219 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 8 |
Hb_002119_110 |
0.1138612588 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 9 |
Hb_000963_120 |
0.1181877665 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001235_120 |
0.1205283407 |
- |
- |
hypothetical protein JCGZ_03395 [Jatropha curcas] |
| 11 |
Hb_004032_190 |
0.1215983551 |
- |
- |
PREDICTED: probable cellulose synthase A catalytic subunit 3 [UDP-forming] isoform X1 [Jatropha curcas] |
| 12 |
Hb_005000_150 |
0.1239920365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000005_010 |
0.1249210161 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 14 |
Hb_186765_010 |
0.1262826395 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 15 |
Hb_003152_020 |
0.1273399364 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 16 |
Hb_080797_010 |
0.1286350426 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 17 |
Hb_001688_050 |
0.1289647558 |
- |
- |
PREDICTED: reticulon-like protein B9 [Jatropha curcas] |
| 18 |
Hb_121624_010 |
0.1313735571 |
- |
- |
PREDICTED: uncharacterized protein LOC105634410 [Jatropha curcas] |
| 19 |
Hb_000103_130 |
0.1326985587 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 20 |
Hb_004374_030 |
0.1329447639 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |