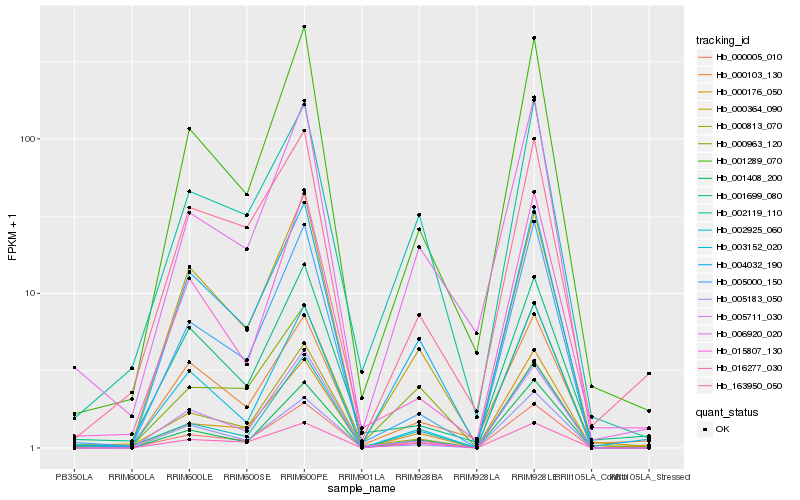
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000005_010 |
0.0 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 2 |
Hb_000176_050 |
0.096021976 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 3 |
Hb_004032_190 |
0.0995716285 |
- |
- |
PREDICTED: probable cellulose synthase A catalytic subunit 3 [UDP-forming] isoform X1 [Jatropha curcas] |
| 4 |
Hb_001289_070 |
0.108164774 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 5 |
Hb_163950_050 |
0.1105193917 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_002119_110 |
0.1150002959 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 7 |
Hb_002925_060 |
0.1153115742 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 8 |
Hb_000813_070 |
0.1166535896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000364_090 |
0.1193379536 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000963_120 |
0.1229126722 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000103_130 |
0.1233515228 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 12 |
Hb_005711_030 |
0.1236383734 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 13 |
Hb_015807_130 |
0.124775952 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006920_020 |
0.1249210161 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 15 |
Hb_001408_200 |
0.1252577589 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 16 |
Hb_005000_150 |
0.1253764783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_016277_030 |
0.125798001 |
- |
- |
hypothetical protein JCGZ_16973 [Jatropha curcas] |
| 18 |
Hb_005183_050 |
0.1278090529 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 19 |
Hb_003152_020 |
0.127966177 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 20 |
Hb_001699_080 |
0.1317967261 |
- |
- |
unnamed protein product [Coffea canephora] |