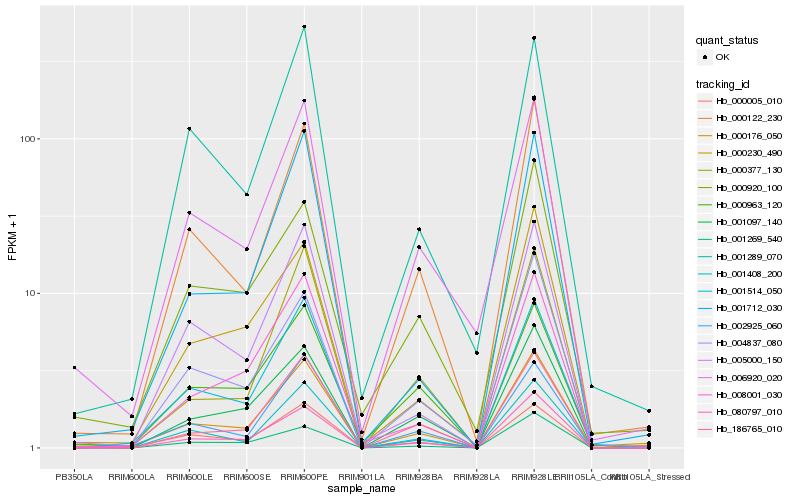
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000176_050 |
0.0 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 2 |
Hb_000122_230 |
0.085829219 |
- |
- |
laccase, putative [Ricinus communis] |
| 3 |
Hb_080797_010 |
0.09119494 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 4 |
Hb_000005_010 |
0.096021976 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 5 |
Hb_006920_020 |
0.0982294667 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 6 |
Hb_001408_200 |
0.1044337942 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 7 |
Hb_001097_140 |
0.1061971251 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002925_060 |
0.1079791457 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 9 |
Hb_005000_150 |
0.1101580743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004837_080 |
0.1111596202 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 11 |
Hb_000230_490 |
0.1115475142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_186765_010 |
0.1121757256 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 13 |
Hb_008001_030 |
0.1157163273 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000920_100 |
0.120067942 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001712_030 |
0.1206721356 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 19 [Jatropha curcas] |
| 16 |
Hb_000377_130 |
0.1217309872 |
- |
- |
hypothetical protein POPTR_0016s00790g [Populus trichocarpa] |
| 17 |
Hb_001269_540 |
0.1219078723 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 18 |
Hb_001514_050 |
0.1223670717 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 19 |
Hb_001289_070 |
0.1233321116 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 20 |
Hb_000963_120 |
0.1235825234 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |