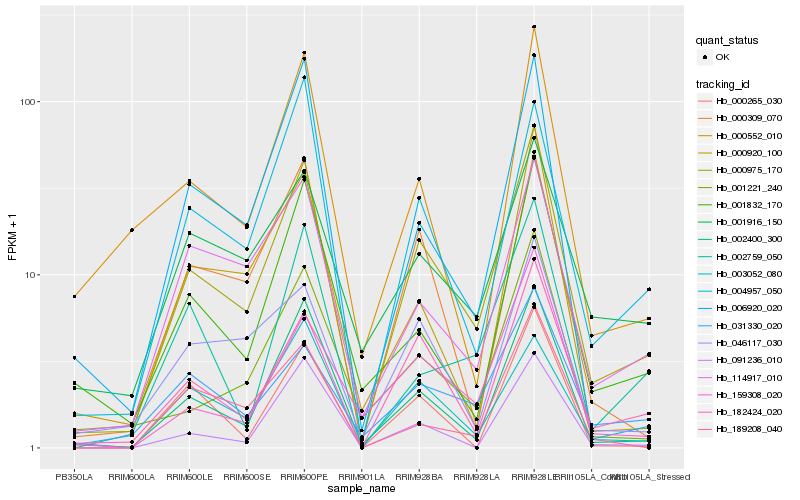
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000975_170 |
0.0 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002400_300 |
0.1170231931 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 3 |
Hb_182424_020 |
0.1207083835 |
- |
- |
hypothetical protein POPTR_0006s24190g [Populus trichocarpa] |
| 4 |
Hb_000309_070 |
0.1541741675 |
- |
- |
PREDICTED: 2-methylene-furan-3-one reductase [Cucumis melo] |
| 5 |
Hb_114917_010 |
0.1559842655 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001221_240 |
0.1575753901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_031330_020 |
0.1604499454 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 8 |
Hb_159308_020 |
0.1607815985 |
- |
- |
hypothetical protein B456_002G027900 [Gossypium raimondii] |
| 9 |
Hb_004957_050 |
0.1609576934 |
- |
- |
hypothetical protein POPTR_0010s12360g [Populus trichocarpa] |
| 10 |
Hb_001832_170 |
0.1613057945 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 11 |
Hb_046117_030 |
0.1619322461 |
- |
- |
sialyltransferase, putative [Ricinus communis] |
| 12 |
Hb_003052_080 |
0.166442281 |
desease resistance |
Gene Name: PRK |
PREDICTED: uridine-cytidine kinase C [Jatropha curcas] |
| 13 |
Hb_006920_020 |
0.1673674673 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 14 |
Hb_001916_150 |
0.1679321092 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Vitis vinifera] |
| 15 |
Hb_000552_010 |
0.167977176 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 2 [Jatropha curcas] |
| 16 |
Hb_000920_100 |
0.169959506 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_091236_010 |
0.1738933076 |
- |
- |
hypothetical protein POPTR_0019s12290g [Populus trichocarpa] |
| 18 |
Hb_189208_040 |
0.1765438786 |
- |
- |
hypothetical protein POPTR_0002s18920g [Populus trichocarpa] |
| 19 |
Hb_000265_030 |
0.1779407364 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 20 |
Hb_002759_050 |
0.178212252 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4 [Jatropha curcas] |