| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
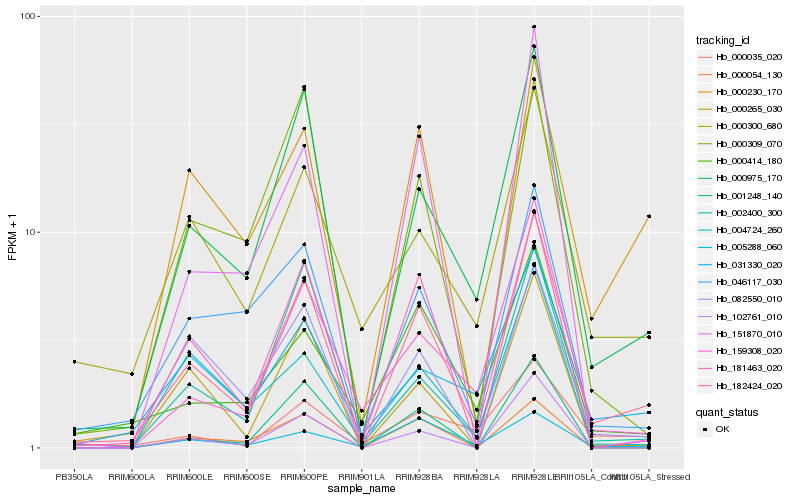
Hb_182424_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s24190g [Populus trichocarpa] |
| 2 |
Hb_000975_170 |
0.1207083835 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_181463_020 |
0.1540269659 |
- |
- |
hypothetical protein JCGZ_26765 [Jatropha curcas] |
| 4 |
Hb_000230_170 |
0.1645566321 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 5 |
Hb_046117_030 |
0.1714415656 |
- |
- |
sialyltransferase, putative [Ricinus communis] |
| 6 |
Hb_031330_020 |
0.1799369109 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 7 |
Hb_082550_010 |
0.1801219946 |
- |
- |
hypothetical protein CISIN_1g0081081mg, partial [Citrus sinensis] |
| 8 |
Hb_159308_020 |
0.1802081381 |
- |
- |
hypothetical protein B456_002G027900 [Gossypium raimondii] |
| 9 |
Hb_000265_030 |
0.182372032 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 10 |
Hb_000300_680 |
0.186418998 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002400_300 |
0.1874116159 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 12 |
Hb_102761_010 |
0.1891215279 |
- |
- |
hypothetical protein JCGZ_21614 [Jatropha curcas] |
| 13 |
Hb_000414_180 |
0.1899168903 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 14 |
Hb_000035_020 |
0.1901725735 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 15 |
Hb_151870_010 |
0.19094127 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_001248_140 |
0.1911288815 |
- |
- |
Thylakoid lumenal 19 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000309_070 |
0.1920859964 |
- |
- |
PREDICTED: 2-methylene-furan-3-one reductase [Cucumis melo] |
| 18 |
Hb_000054_130 |
0.1937078143 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 19 |
Hb_005288_060 |
0.1971199572 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 20 |
Hb_004724_260 |
0.1986099297 |
- |
- |
PREDICTED: 40S ribosomal protein S14-2-like [Gossypium raimondii] |