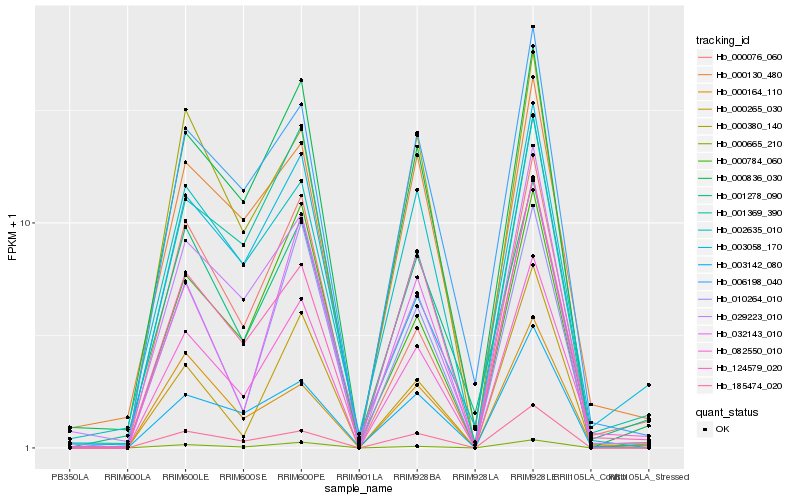
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_082550_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0081081mg, partial [Citrus sinensis] |
| 2 |
Hb_000836_030 |
0.1112975957 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 3 |
Hb_006198_040 |
0.1161607296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000130_480 |
0.1190648202 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 5 |
Hb_000665_210 |
0.1216077302 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 6 |
Hb_124579_020 |
0.1222186634 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 7 |
Hb_000076_060 |
0.1232143554 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 8 |
Hb_010264_010 |
0.1245582995 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR-LIKE 1-like [Jatropha curcas] |
| 9 |
Hb_001278_090 |
0.1263824196 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 10 |
Hb_003142_080 |
0.1264625722 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 11 |
Hb_000784_060 |
0.1296022285 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_029223_010 |
0.1300265934 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 13 |
Hb_001369_390 |
0.1303300277 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 14 |
Hb_185474_020 |
0.1325582936 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 15 |
Hb_000265_030 |
0.1353586444 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 16 |
Hb_000380_140 |
0.1375489892 |
- |
- |
PREDICTED: uncharacterized protein LOC105628069 [Jatropha curcas] |
| 17 |
Hb_000164_110 |
0.1378671128 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 18 |
Hb_002635_010 |
0.1384430955 |
- |
- |
homogentisate phytyl transferase [Hevea brasiliensis] |
| 19 |
Hb_003058_170 |
0.1384813823 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 20 |
Hb_032143_010 |
0.1398173378 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |