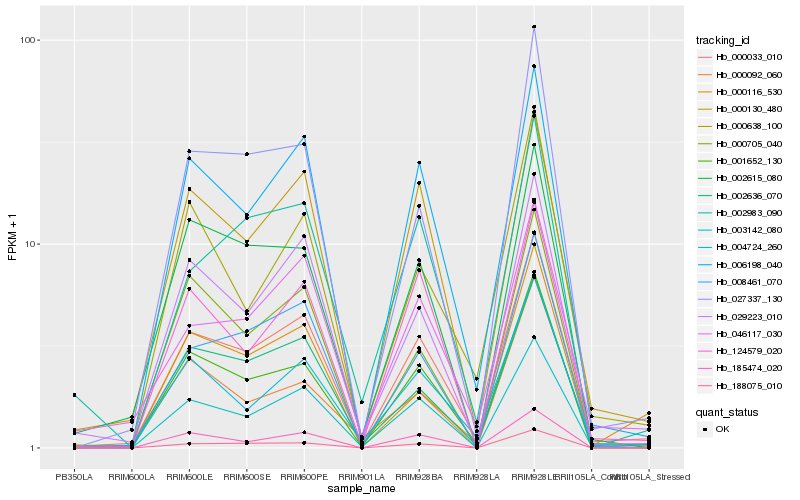
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000033_010 |
0.0 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 2 |
Hb_006198_040 |
0.0806145785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003142_080 |
0.0886508525 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 4 |
Hb_001652_130 |
0.0895789056 |
- |
- |
hypothetical protein B456_002G193900 [Gossypium raimondii] |
| 5 |
Hb_002636_070 |
0.0976336891 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 6 |
Hb_004724_260 |
0.0987848889 |
- |
- |
PREDICTED: 40S ribosomal protein S14-2-like [Gossypium raimondii] |
| 7 |
Hb_029223_010 |
0.1036904015 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 8 |
Hb_027337_130 |
0.1057964346 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 9 |
Hb_008461_070 |
0.1068649543 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000130_480 |
0.1070067756 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 11 |
Hb_188075_010 |
0.1101823508 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 12 |
Hb_185474_020 |
0.113012577 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 13 |
Hb_046117_030 |
0.1145935557 |
- |
- |
sialyltransferase, putative [Ricinus communis] |
| 14 |
Hb_124579_020 |
0.1165336924 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 15 |
Hb_000638_100 |
0.1226327352 |
- |
- |
PREDICTED: histone deacetylase 14 [Jatropha curcas] |
| 16 |
Hb_000705_040 |
0.1250398433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002615_080 |
0.1263033492 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 18 |
Hb_002983_090 |
0.1316482339 |
- |
- |
Altered inheritance of mitochondria protein 32 [Glycine soja] |
| 19 |
Hb_000116_530 |
0.1366628688 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 3; Short=HMG-CoA reductase 3 [Hevea brasiliensis] |
| 20 |
Hb_000092_060 |
0.1381701397 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |