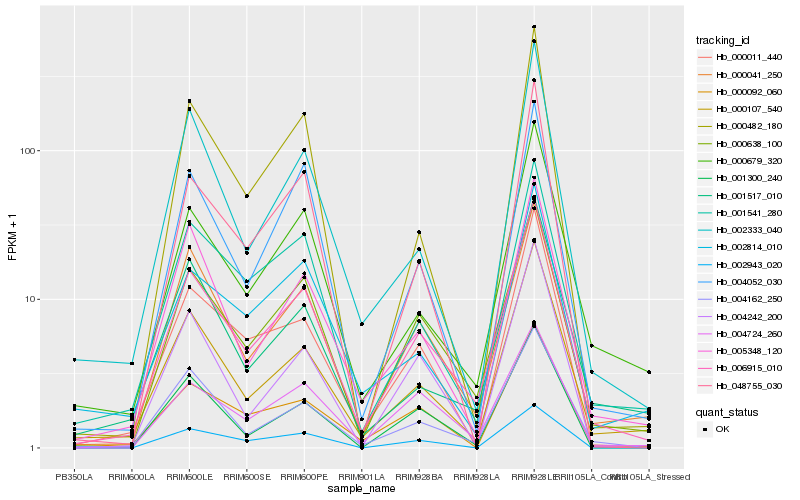
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006915_010 |
0.0 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 2 |
Hb_000638_100 |
0.0590721118 |
- |
- |
PREDICTED: histone deacetylase 14 [Jatropha curcas] |
| 3 |
Hb_000041_250 |
0.0893890236 |
- |
- |
PREDICTED: uncharacterized protein LOC105641253 [Jatropha curcas] |
| 4 |
Hb_004162_250 |
0.095312663 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 5 |
Hb_000107_540 |
0.0973880992 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 6 |
Hb_000679_320 |
0.103647521 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 7 |
Hb_001541_280 |
0.104354516 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 8 |
Hb_004724_260 |
0.1072045755 |
- |
- |
PREDICTED: 40S ribosomal protein S14-2-like [Gossypium raimondii] |
| 9 |
Hb_000092_060 |
0.11126423 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 10 |
Hb_000011_440 |
0.1147818358 |
- |
- |
PREDICTED: GDSL esterase/lipase 1 [Populus euphratica] |
| 11 |
Hb_004052_030 |
0.115920277 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 12 |
Hb_005348_120 |
0.1165588861 |
- |
- |
PREDICTED: chlorophyll a-b binding protein, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001300_240 |
0.1220310567 |
- |
- |
shikimate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_001517_010 |
0.1268586381 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002943_020 |
0.1268945067 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 16 |
Hb_002333_040 |
0.1269393973 |
- |
- |
hypothetical protein CISIN_1g015895mg [Citrus sinensis] |
| 17 |
Hb_004242_200 |
0.1281691142 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 18 |
Hb_000482_180 |
0.1290121328 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 19 |
Hb_048755_030 |
0.1296440836 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 20 |
Hb_002814_010 |
0.1304909056 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |