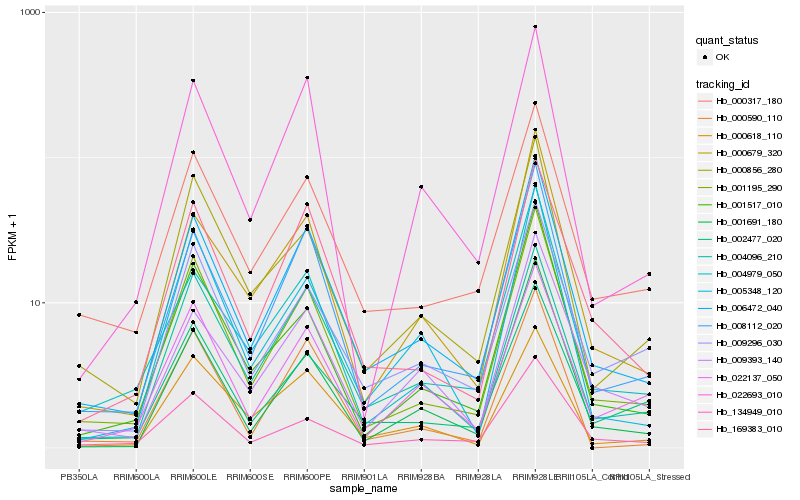
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006472_040 |
0.0 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_008112_020 |
0.0618044682 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001195_290 |
0.0705408628 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 4 |
Hb_000679_320 |
0.091922523 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 5 |
Hb_169383_010 |
0.0979758056 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000317_180 |
0.1011230647 |
- |
- |
PREDICTED: protein THYLAKOID FORMATION1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_009393_140 |
0.1063067837 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004979_050 |
0.1069921723 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 9 |
Hb_004096_210 |
0.1071991424 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 10 |
Hb_022693_010 |
0.1084494214 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 11 |
Hb_001517_010 |
0.1094377289 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000618_110 |
0.1103021047 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 13 |
Hb_134949_010 |
0.1131161711 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 14 |
Hb_000856_280 |
0.1131657906 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 15 |
Hb_001691_180 |
0.1139828092 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 16 |
Hb_002477_020 |
0.1146262854 |
- |
- |
PREDICTED: uncharacterized protein LOC105631402 [Jatropha curcas] |
| 17 |
Hb_009296_030 |
0.1157372962 |
- |
- |
PREDICTED: uncharacterized protein LOC105634328 [Jatropha curcas] |
| 18 |
Hb_005348_120 |
0.1190627193 |
- |
- |
PREDICTED: chlorophyll a-b binding protein, chloroplastic [Jatropha curcas] |
| 19 |
Hb_022137_050 |
0.1196801136 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 20 |
Hb_000590_110 |
0.1228905874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |