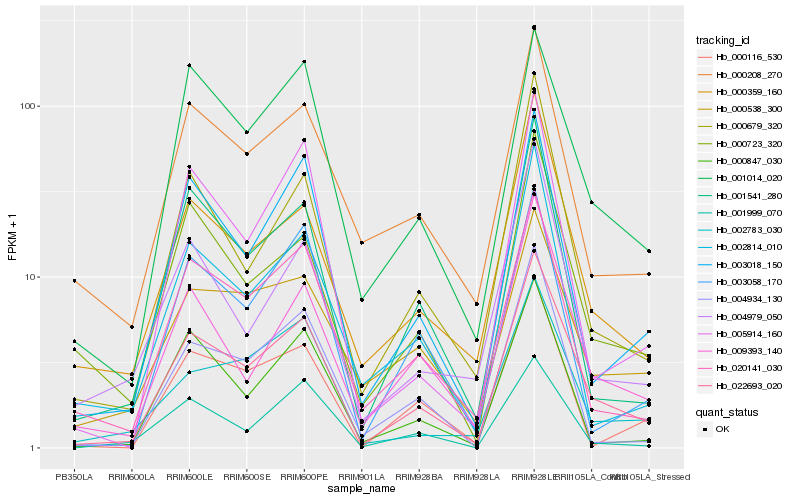
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022693_020 |
0.0 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 2 |
Hb_000679_320 |
0.1191891231 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 3 |
Hb_003018_150 |
0.1218624868 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 4 |
Hb_009393_140 |
0.1222243763 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000359_160 |
0.1227776273 |
- |
- |
PREDICTED: protein CAJ1 [Jatropha curcas] |
| 6 |
Hb_001541_280 |
0.1234138919 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 7 |
Hb_020141_030 |
0.125281452 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 8 |
Hb_000723_320 |
0.1310004899 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 9 |
Hb_001014_020 |
0.1381213153 |
- |
- |
gene GA family protein [Populus trichocarpa] |
| 10 |
Hb_000208_270 |
0.1399538359 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 11 |
Hb_002814_010 |
0.1400723133 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004934_130 |
0.1419031865 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 13 |
Hb_005914_160 |
0.1448263837 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000847_030 |
0.1456216553 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 15 |
Hb_002783_030 |
0.146385592 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 16 |
Hb_003058_170 |
0.1484498071 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000116_530 |
0.1502838269 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 3; Short=HMG-CoA reductase 3 [Hevea brasiliensis] |
| 18 |
Hb_004979_050 |
0.1504249359 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 19 |
Hb_000538_300 |
0.1529543975 |
- |
- |
PREDICTED: WAT1-related protein At3g02690, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001999_070 |
0.1531843269 |
- |
- |
PREDICTED: ammonium transporter 1 member 3 [Jatropha curcas] |