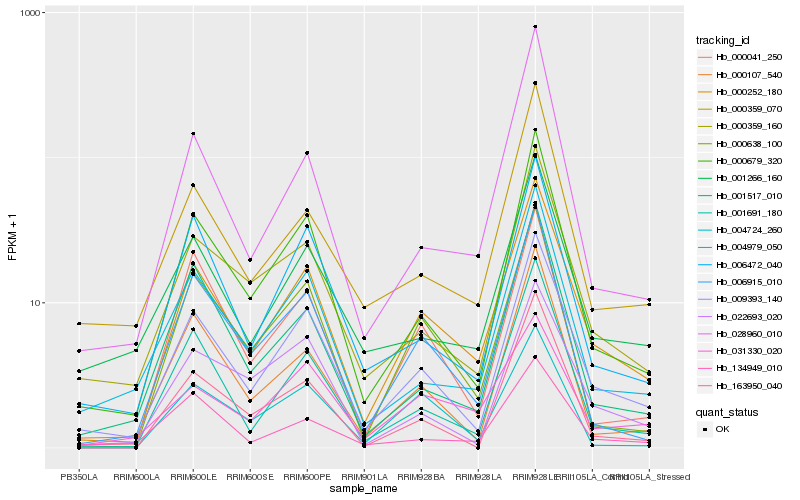
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000252_180 |
0.0 |
- |
- |
Glutamate--glyoxylate aminotransferase 2 [Morus notabilis] |
| 2 |
Hb_009393_140 |
0.0871870967 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000679_320 |
0.1189296237 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 4 |
Hb_006915_010 |
0.1305418962 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 5 |
Hb_000638_100 |
0.1313936505 |
- |
- |
PREDICTED: histone deacetylase 14 [Jatropha curcas] |
| 6 |
Hb_001691_180 |
0.1343557027 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 7 |
Hb_000359_160 |
0.1391881683 |
- |
- |
PREDICTED: protein CAJ1 [Jatropha curcas] |
| 8 |
Hb_031330_020 |
0.1423153858 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 9 |
Hb_004979_050 |
0.1455266216 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 10 |
Hb_006472_040 |
0.1464307035 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001266_160 |
0.1509095075 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000107_540 |
0.1516358225 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 13 |
Hb_028960_010 |
0.1528135028 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 14 |
Hb_163950_040 |
0.1546546451 |
- |
- |
PREDICTED: putative 4-hydroxy-tetrahydrodipicolinate reductase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000359_070 |
0.1562972128 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000041_250 |
0.1567124601 |
- |
- |
PREDICTED: uncharacterized protein LOC105641253 [Jatropha curcas] |
| 17 |
Hb_001517_010 |
0.1573036586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004724_260 |
0.1605516102 |
- |
- |
PREDICTED: 40S ribosomal protein S14-2-like [Gossypium raimondii] |
| 19 |
Hb_022693_020 |
0.1609090228 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 20 |
Hb_134949_010 |
0.1610191602 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |