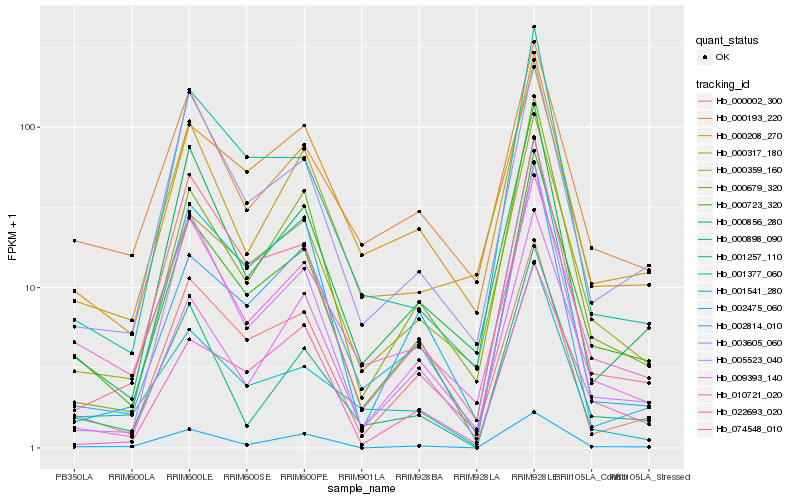
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000723_320 |
0.0 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 2 |
Hb_000359_160 |
0.1077591076 |
- |
- |
PREDICTED: protein CAJ1 [Jatropha curcas] |
| 3 |
Hb_010721_020 |
0.1148017489 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002475_060 |
0.118015043 |
- |
- |
PREDICTED: potassium channel SKOR-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_003605_060 |
0.1192690892 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000208_270 |
0.1234244916 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 7 |
Hb_009393_140 |
0.1250303304 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000679_320 |
0.1257990693 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 9 |
Hb_005523_040 |
0.126273468 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 10 |
Hb_000002_300 |
0.1270843866 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 11 |
Hb_000898_090 |
0.1281253033 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105633600 isoform X1 [Jatropha curcas] |
| 12 |
Hb_074548_010 |
0.1286157206 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_000856_280 |
0.1295151819 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 14 |
Hb_022693_020 |
0.1310004899 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 15 |
Hb_001541_280 |
0.1317301326 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 16 |
Hb_000193_220 |
0.1322427878 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001377_060 |
0.1324165621 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01030, mitochondrial [Jatropha curcas] |
| 18 |
Hb_002814_010 |
0.1331155266 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001257_110 |
0.1363886974 |
- |
- |
PREDICTED: uncharacterized protein LOC105636396 [Jatropha curcas] |
| 20 |
Hb_000317_180 |
0.1368437673 |
- |
- |
PREDICTED: protein THYLAKOID FORMATION1, chloroplastic [Jatropha curcas] |