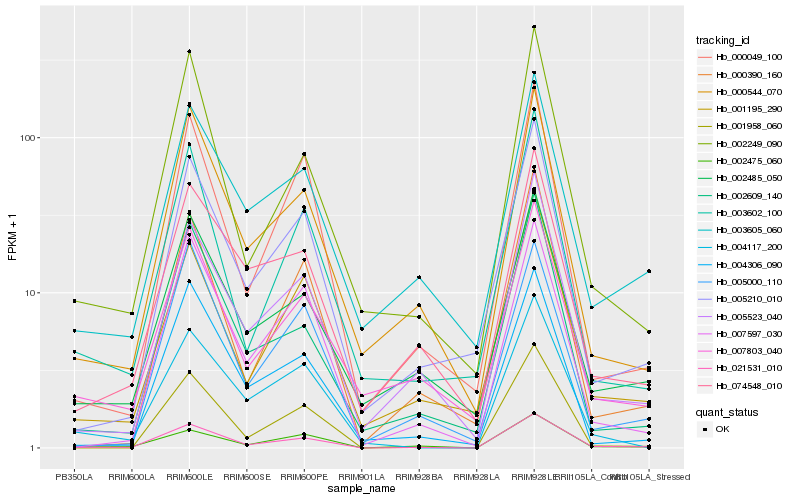
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021531_010 |
0.0 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 2 |
Hb_005523_040 |
0.104886688 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 3 |
Hb_000544_070 |
0.1059774495 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |
| 4 |
Hb_007597_030 |
0.1090314518 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_005210_010 |
0.1137351888 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 6 |
Hb_002475_060 |
0.1155968624 |
- |
- |
PREDICTED: potassium channel SKOR-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002249_090 |
0.1181853921 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002485_050 |
0.118626496 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000049_100 |
0.1186709251 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 10 |
Hb_001195_290 |
0.1201491575 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 11 |
Hb_001958_060 |
0.1204118656 |
- |
- |
PREDICTED: calcium uptake protein 1, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_074548_010 |
0.12109907 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_003605_060 |
0.1215748606 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003602_100 |
0.1217436887 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_004306_090 |
0.1247691788 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007803_040 |
0.1268075834 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002609_140 |
0.1276288316 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_004117_200 |
0.1278110892 |
- |
- |
PREDICTED: RING-H2 finger protein ATL74 [Jatropha curcas] |
| 19 |
Hb_000390_160 |
0.1298755048 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 20 |
Hb_005000_110 |
0.1314712409 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |