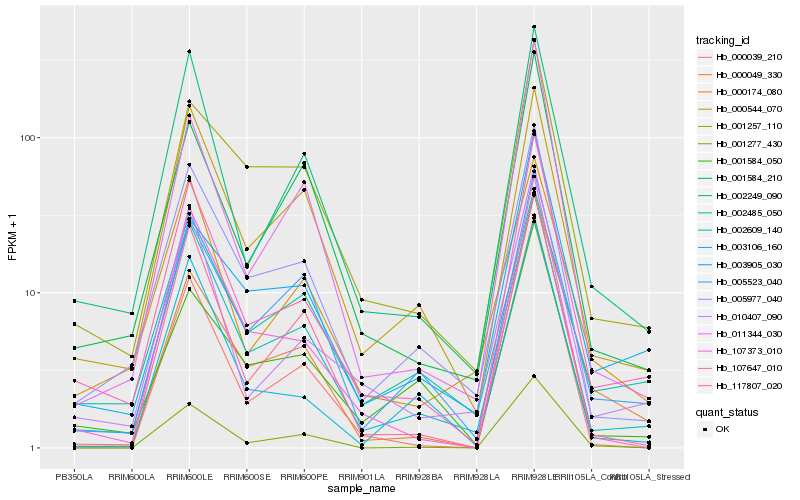
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_107647_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein ycf39 [Jatropha curcas] |
| 2 |
Hb_002609_140 |
0.088264581 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_010407_090 |
0.0942405935 |
- |
- |
PREDICTED: aldo-keto reductase isoform X1 [Jatropha curcas] |
| 4 |
Hb_002249_090 |
0.1032290004 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005977_040 |
0.1146329871 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |
| 6 |
Hb_107373_010 |
0.1191742806 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_117807_020 |
0.1312953034 |
- |
- |
methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_011344_030 |
0.1324081494 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000049_330 |
0.1332508036 |
- |
- |
PREDICTED: phospholipase A1-IIdelta [Jatropha curcas] |
| 10 |
Hb_005523_040 |
0.13359013 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 11 |
Hb_003106_160 |
0.13487762 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 12 |
Hb_001277_430 |
0.136701105 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 13 |
Hb_000544_070 |
0.1373098748 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |
| 14 |
Hb_000174_080 |
0.1376667921 |
- |
- |
4-nitrophenylphosphatase, putative [Ricinus communis] |
| 15 |
Hb_001584_210 |
0.1382714461 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002485_050 |
0.1399724757 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001257_110 |
0.1406100186 |
- |
- |
PREDICTED: uncharacterized protein LOC105636396 [Jatropha curcas] |
| 18 |
Hb_003905_030 |
0.1409605156 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 19 |
Hb_000039_210 |
0.1422828787 |
- |
- |
PREDICTED: uncharacterized protein LOC105649449 [Jatropha curcas] |
| 20 |
Hb_001584_050 |
0.1423012198 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X1 [Jatropha curcas] |