| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
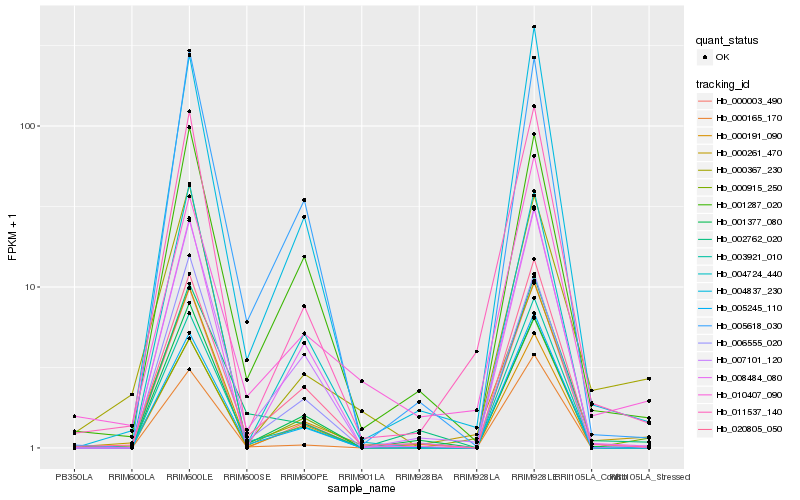
Hb_000261_470 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 2 |
Hb_011537_140 |
0.0620095983 |
- |
- |
hypothetical protein POPTR_0001s05320g [Populus trichocarpa] |
| 3 |
Hb_000367_230 |
0.1182675076 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_020805_050 |
0.1227744832 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 5 |
Hb_007101_120 |
0.1253158028 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 6 |
Hb_004837_230 |
0.1290054327 |
- |
- |
unknown [Populus trichocarpa] |
| 7 |
Hb_001287_020 |
0.1352410076 |
- |
- |
SulA [Manihot esculenta] |
| 8 |
Hb_003921_010 |
0.1374206033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010407_090 |
0.1379753931 |
- |
- |
PREDICTED: aldo-keto reductase isoform X1 [Jatropha curcas] |
| 10 |
Hb_000003_490 |
0.1382730365 |
- |
- |
PREDICTED: uncharacterized protein LOC105631559 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000915_250 |
0.1395153137 |
- |
- |
chloroplast latex aldolase-like protein [Manihot esculenta] |
| 12 |
Hb_000191_090 |
0.142382785 |
transcription factor |
TF Family: MYB |
GHMYB10, putative [Theobroma cacao] |
| 13 |
Hb_005245_110 |
0.1437445442 |
- |
- |
hypothetical protein CISIN_1g0225141mg [Citrus sinensis] |
| 14 |
Hb_006555_020 |
0.1437556686 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 15 |
Hb_002762_020 |
0.1449232839 |
- |
- |
hypothetical protein VITISV_043202 [Vitis vinifera] |
| 16 |
Hb_005618_030 |
0.1449982735 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 17 |
Hb_008484_080 |
0.1452264056 |
- |
- |
PREDICTED: crocetin glucosyltransferase, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_004724_440 |
0.1469484451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001377_080 |
0.1474143012 |
- |
- |
PREDICTED: uncharacterized protein LOC105642864 [Jatropha curcas] |
| 20 |
Hb_000165_170 |
0.1509938882 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |