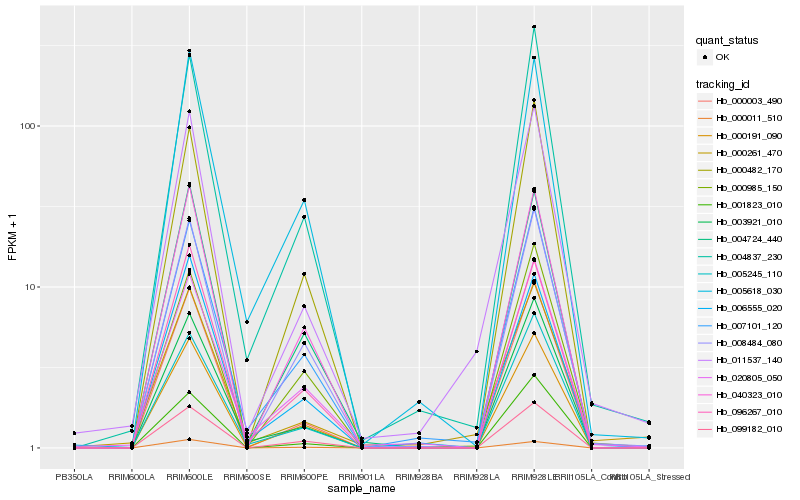
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011537_140 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s05320g [Populus trichocarpa] |
| 2 |
Hb_000261_470 |
0.0620095983 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 3 |
Hb_007101_120 |
0.1027277861 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 4 |
Hb_000003_490 |
0.10281296 |
- |
- |
PREDICTED: uncharacterized protein LOC105631559 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004837_230 |
0.108815396 |
- |
- |
unknown [Populus trichocarpa] |
| 6 |
Hb_000191_090 |
0.1111105148 |
transcription factor |
TF Family: MYB |
GHMYB10, putative [Theobroma cacao] |
| 7 |
Hb_020805_050 |
0.1131029575 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 8 |
Hb_003921_010 |
0.1146831889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_008484_080 |
0.1156707561 |
- |
- |
PREDICTED: crocetin glucosyltransferase, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_004724_440 |
0.1181141116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005245_110 |
0.1195677175 |
- |
- |
hypothetical protein CISIN_1g0225141mg [Citrus sinensis] |
| 12 |
Hb_040323_010 |
0.1220491329 |
- |
- |
- |
| 13 |
Hb_099182_010 |
0.1221042196 |
- |
- |
PREDICTED: uncharacterized protein LOC101214783 [Cucumis sativus] |
| 14 |
Hb_001823_010 |
0.1244546023 |
- |
- |
PREDICTED: ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 15 |
Hb_096267_010 |
0.1258566034 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 16 |
Hb_006555_020 |
0.1279409473 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 17 |
Hb_005618_030 |
0.1284250317 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 18 |
Hb_000482_170 |
0.1284621598 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |
| 19 |
Hb_000985_150 |
0.1292231431 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 20 |
Hb_000011_510 |
0.1308213164 |
- |
- |
Esterase precursor, putative [Ricinus communis] |