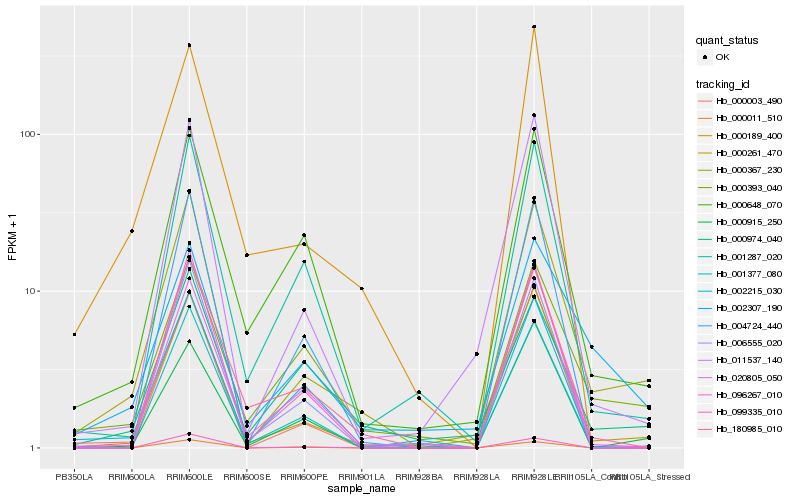
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_230 |
0.0 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000261_470 |
0.1182675076 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 3 |
Hb_011537_140 |
0.1504688583 |
- |
- |
hypothetical protein POPTR_0001s05320g [Populus trichocarpa] |
| 4 |
Hb_180985_010 |
0.1712426226 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 1 [Theobroma cacao] |
| 5 |
Hb_000393_040 |
0.1720724405 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 6 |
Hb_002307_190 |
0.1724344906 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |
| 7 |
Hb_000648_070 |
0.1746606978 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 8 |
Hb_001287_020 |
0.1759387085 |
- |
- |
SulA [Manihot esculenta] |
| 9 |
Hb_000974_040 |
0.1794495036 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 10 |
Hb_004724_440 |
0.180834598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006555_020 |
0.1811371357 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 12 |
Hb_000915_250 |
0.1816291908 |
- |
- |
chloroplast latex aldolase-like protein [Manihot esculenta] |
| 13 |
Hb_002215_030 |
0.1830973079 |
- |
- |
PREDICTED: uncharacterized protein LOC105640840 isoform X1 [Jatropha curcas] |
| 14 |
Hb_020805_050 |
0.183314325 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 15 |
Hb_096267_010 |
0.1834911487 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 16 |
Hb_000011_510 |
0.183768607 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 17 |
Hb_000003_490 |
0.1860243928 |
- |
- |
PREDICTED: uncharacterized protein LOC105631559 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001377_080 |
0.1864282947 |
- |
- |
PREDICTED: uncharacterized protein LOC105642864 [Jatropha curcas] |
| 19 |
Hb_000189_400 |
0.1867788729 |
- |
- |
hypothetical protein CathTA2_0066 [Caldalkalibacillus thermarum TA2.A1] |
| 20 |
Hb_099335_010 |
0.1882886303 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C2-like [Populus euphratica] |