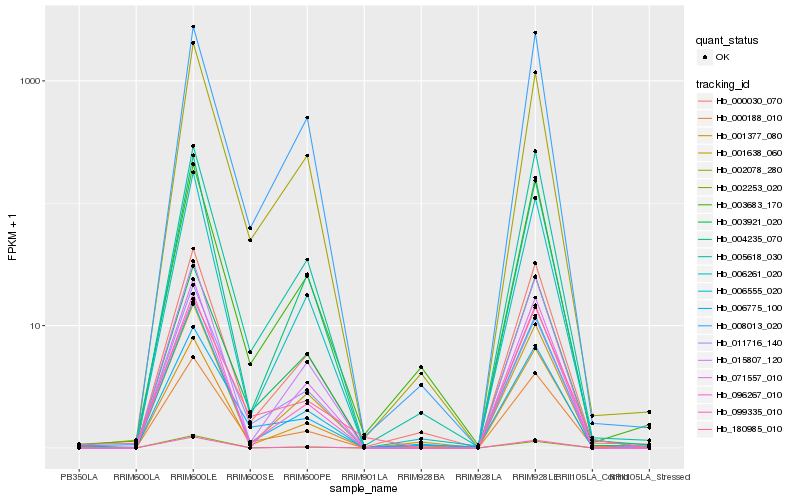
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000188_010 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 94C1-like [Jatropha curcas] |
| 2 |
Hb_002078_280 |
0.0746199907 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 3 |
Hb_000030_070 |
0.0759557802 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006261_020 |
0.0775695851 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 5 |
Hb_003921_020 |
0.081560935 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 6 |
Hb_004235_070 |
0.0819821533 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 7 |
Hb_096267_010 |
0.082209441 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 8 |
Hb_011716_140 |
0.0826751586 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 9 |
Hb_005618_030 |
0.0837732511 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 10 |
Hb_006555_020 |
0.0864295674 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 11 |
Hb_002253_020 |
0.0876233824 |
- |
- |
PREDICTED: callose synthase 5 [Jatropha curcas] |
| 12 |
Hb_006775_100 |
0.0896153743 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_015807_120 |
0.0914997882 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_180985_010 |
0.0946387795 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 1 [Theobroma cacao] |
| 15 |
Hb_001377_080 |
0.0961691887 |
- |
- |
PREDICTED: uncharacterized protein LOC105642864 [Jatropha curcas] |
| 16 |
Hb_001638_060 |
0.0964309755 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 17 |
Hb_003683_170 |
0.0982513217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_099335_010 |
0.1009826753 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C2-like [Populus euphratica] |
| 19 |
Hb_008013_020 |
0.1014168223 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 20 |
Hb_071557_010 |
0.1029696027 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |