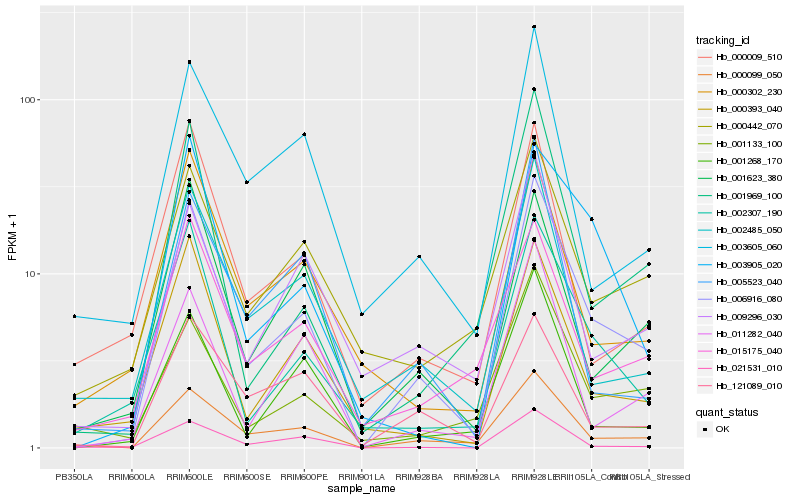
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006916_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 2 |
Hb_000099_050 |
0.1303033732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002307_190 |
0.1526734086 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |
| 4 |
Hb_000393_040 |
0.1670806678 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 5 |
Hb_021531_010 |
0.1691938366 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 6 |
Hb_001969_100 |
0.1698184955 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 7 |
Hb_002485_050 |
0.1698598148 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001268_170 |
0.1713338072 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 9 |
Hb_001623_380 |
0.1728279045 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_011282_040 |
0.1739737148 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 11 |
Hb_003905_020 |
0.1763798735 |
- |
- |
PREDICTED: uncharacterized protein LOC105131878 isoform X1 [Populus euphratica] |
| 12 |
Hb_009296_030 |
0.1767432969 |
- |
- |
PREDICTED: uncharacterized protein LOC105634328 [Jatropha curcas] |
| 13 |
Hb_015175_040 |
0.1793626778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003605_060 |
0.1812348449 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 15 |
Hb_121089_010 |
0.1826508815 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 16 |
Hb_001133_100 |
0.1831835435 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 17 |
Hb_000302_230 |
0.1832409986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005523_040 |
0.1837768958 |
- |
- |
PREDICTED: uncharacterized protein LOC100253680 [Vitis vinifera] |
| 19 |
Hb_000009_510 |
0.1845328641 |
- |
- |
PREDICTED: uncharacterized protein LOC105126496 [Populus euphratica] |
| 20 |
Hb_000442_070 |
0.1846186582 |
- |
- |
PREDICTED: uncharacterized protein LOC105645538 [Jatropha curcas] |