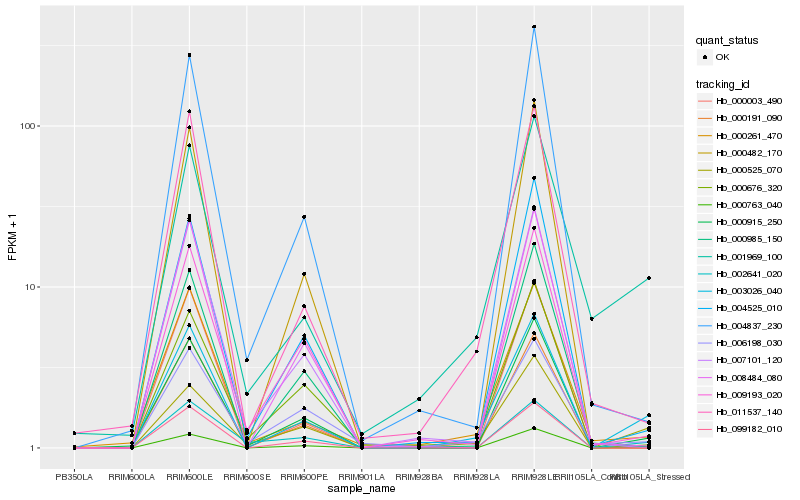
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003026_040 |
0.0 |
- |
- |
Uncharacterized protein TCM_007861 [Theobroma cacao] |
| 2 |
Hb_000763_040 |
0.1020433915 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 3 |
Hb_000525_070 |
0.1356529164 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like [Jatropha curcas] |
| 4 |
Hb_000915_250 |
0.1361082748 |
- |
- |
chloroplast latex aldolase-like protein [Manihot esculenta] |
| 5 |
Hb_001969_100 |
0.1500440517 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 6 |
Hb_000261_470 |
0.1537124169 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 7 |
Hb_011537_140 |
0.1657796619 |
- |
- |
hypothetical protein POPTR_0001s05320g [Populus trichocarpa] |
| 8 |
Hb_002641_020 |
0.1710457862 |
- |
- |
3'-5' exonuclease, putative [Ricinus communis] |
| 9 |
Hb_004525_010 |
0.1722912719 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 10 |
Hb_006198_030 |
0.1772006017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_009193_020 |
0.1790345771 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 12 |
Hb_008484_080 |
0.1812044403 |
- |
- |
PREDICTED: crocetin glucosyltransferase, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_099182_010 |
0.1827232229 |
- |
- |
PREDICTED: uncharacterized protein LOC101214783 [Cucumis sativus] |
| 14 |
Hb_000985_150 |
0.1833667666 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 15 |
Hb_000191_090 |
0.1834033181 |
transcription factor |
TF Family: MYB |
GHMYB10, putative [Theobroma cacao] |
| 16 |
Hb_007101_120 |
0.1845676724 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 17 |
Hb_004837_230 |
0.1848976334 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_000482_170 |
0.1850245033 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |
| 19 |
Hb_000003_490 |
0.1874745064 |
- |
- |
PREDICTED: uncharacterized protein LOC105631559 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000676_320 |
0.1877464289 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |