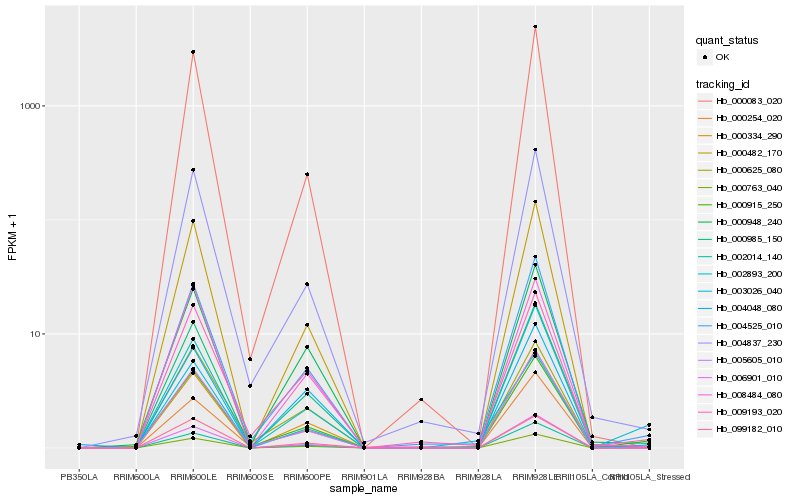
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000763_040 |
0.0 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 2 |
Hb_000915_250 |
0.0718199842 |
- |
- |
chloroplast latex aldolase-like protein [Manihot esculenta] |
| 3 |
Hb_003026_040 |
0.1020433915 |
- |
- |
Uncharacterized protein TCM_007861 [Theobroma cacao] |
| 4 |
Hb_004525_010 |
0.1136248209 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 5 |
Hb_000334_290 |
0.1222898696 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 6 |
Hb_000254_020 |
0.1244803697 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 7 |
Hb_000985_150 |
0.1299814111 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 8 |
Hb_000482_170 |
0.1301026534 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |
| 9 |
Hb_009193_020 |
0.1323986904 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 10 |
Hb_006901_010 |
0.134464675 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 11 |
Hb_008484_080 |
0.135652326 |
- |
- |
PREDICTED: crocetin glucosyltransferase, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000948_240 |
0.1380220285 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002014_140 |
0.1382206097 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 14 |
Hb_004837_230 |
0.1384516712 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_005605_010 |
0.138690219 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 16 |
Hb_099182_010 |
0.1391475113 |
- |
- |
PREDICTED: uncharacterized protein LOC101214783 [Cucumis sativus] |
| 17 |
Hb_002893_200 |
0.1408742636 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_000083_020 |
0.141909451 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 19 |
Hb_004048_080 |
0.142207304 |
- |
- |
hypothetical protein POPTR_0001s40360g [Populus trichocarpa] |
| 20 |
Hb_000625_080 |
0.1425481997 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |