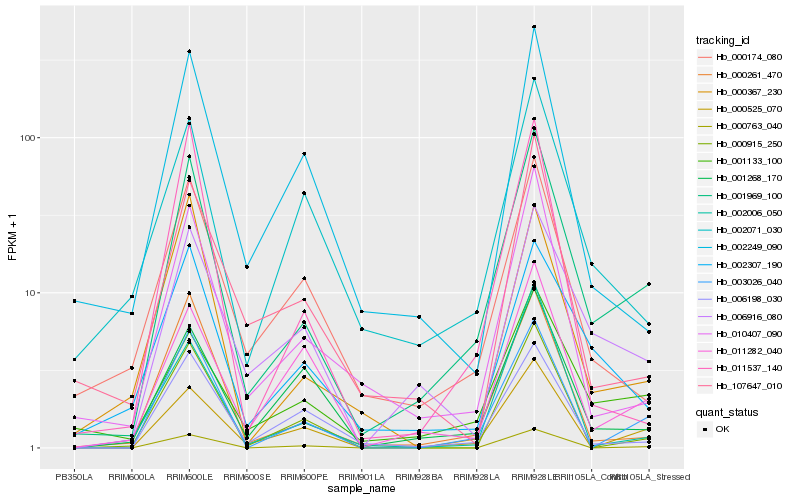
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001969_100 |
0.0 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 2 |
Hb_003026_040 |
0.1500440517 |
- |
- |
Uncharacterized protein TCM_007861 [Theobroma cacao] |
| 3 |
Hb_000261_470 |
0.1528746058 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 4 |
Hb_000525_070 |
0.160209713 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like [Jatropha curcas] |
| 5 |
Hb_001133_100 |
0.1622626141 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 6 |
Hb_010407_090 |
0.1641611748 |
- |
- |
PREDICTED: aldo-keto reductase isoform X1 [Jatropha curcas] |
| 7 |
Hb_006916_080 |
0.1698184955 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 8 |
Hb_000763_040 |
0.1819284036 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 9 |
Hb_002307_190 |
0.1826741873 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |
| 10 |
Hb_006198_030 |
0.186392516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_011537_140 |
0.1883893938 |
- |
- |
hypothetical protein POPTR_0001s05320g [Populus trichocarpa] |
| 12 |
Hb_000367_230 |
0.1886722036 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000174_080 |
0.1897002988 |
- |
- |
4-nitrophenylphosphatase, putative [Ricinus communis] |
| 14 |
Hb_000915_250 |
0.1902400906 |
- |
- |
chloroplast latex aldolase-like protein [Manihot esculenta] |
| 15 |
Hb_001268_170 |
0.1919645163 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 16 |
Hb_107647_010 |
0.1935175474 |
- |
- |
PREDICTED: uncharacterized protein ycf39 [Jatropha curcas] |
| 17 |
Hb_011282_040 |
0.1939526744 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 18 |
Hb_002071_030 |
0.1941613102 |
- |
- |
PREDICTED: ACT domain-containing protein ACR11-like isoform X1 [Gossypium raimondii] |
| 19 |
Hb_002006_050 |
0.1969023011 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 20 |
Hb_002249_090 |
0.1982057757 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |