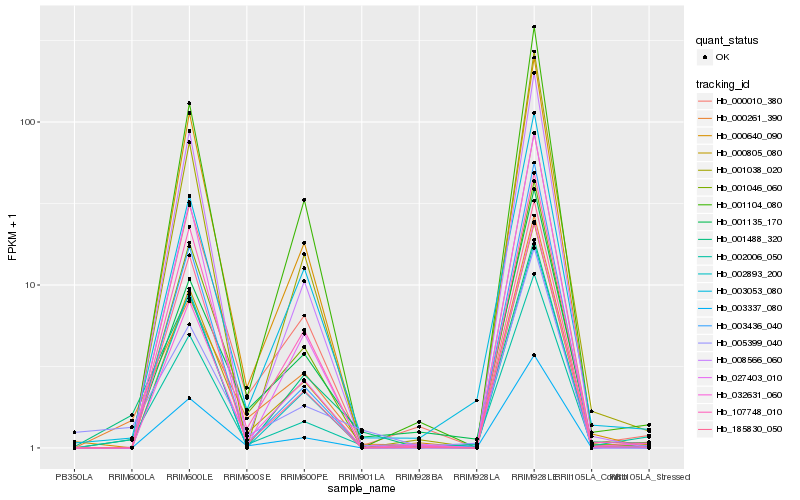
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002006_050 |
0.0 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 2 |
Hb_000261_390 |
0.1094549915 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 3 |
Hb_000805_080 |
0.1104376397 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000010_380 |
0.1112845382 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 5 |
Hb_032631_060 |
0.1129939063 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |
| 6 |
Hb_185830_050 |
0.1151703248 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Jatropha curcas] |
| 7 |
Hb_003053_080 |
0.1190711804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_008566_060 |
0.1202298022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001135_170 |
0.1206143997 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 10 |
Hb_003337_080 |
0.1219003701 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 11 |
Hb_001046_060 |
0.1226088765 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |
| 12 |
Hb_003436_040 |
0.1228508394 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 13 |
Hb_027403_010 |
0.1232954825 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 14 |
Hb_001104_080 |
0.123481732 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 15 |
Hb_001488_320 |
0.123753529 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 16 |
Hb_000640_090 |
0.1249401151 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_002893_200 |
0.1249827161 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_005399_040 |
0.1250686892 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 19 |
Hb_107748_010 |
0.1250795588 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_001038_020 |
0.1255267037 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |