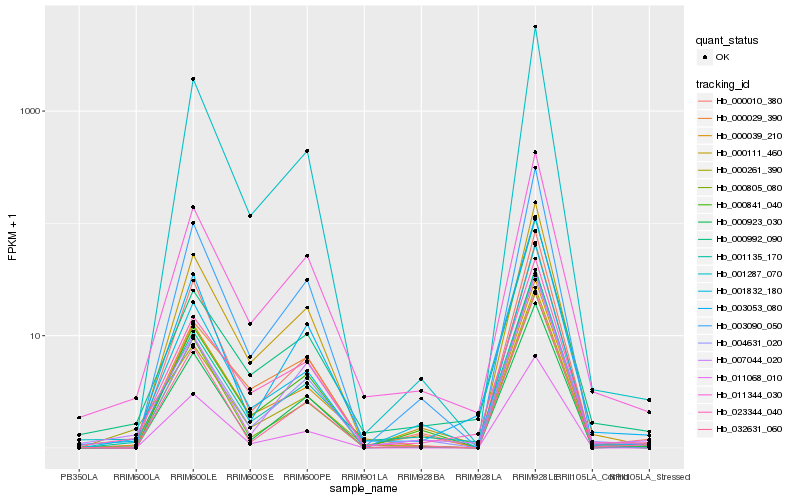
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_390 |
0.0 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 2 |
Hb_001135_170 |
0.0758806924 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 3 |
Hb_001832_180 |
0.0845168388 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 4 |
Hb_011344_030 |
0.0905707356 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_023344_040 |
0.0929625916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004631_020 |
0.095892584 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 7 |
Hb_001287_070 |
0.0959556656 |
- |
- |
latex plastidic aldolase-like protein [Hevea brasiliensis] |
| 8 |
Hb_000010_380 |
0.0980148996 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 9 |
Hb_000992_090 |
0.0988324142 |
transcription factor |
TF Family: Orphans |
CIL, putative [Ricinus communis] |
| 10 |
Hb_032631_060 |
0.0989712843 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |
| 11 |
Hb_003090_050 |
0.09939039 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 12 |
Hb_007044_020 |
0.1018876627 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_000111_460 |
0.1034676155 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 14 |
Hb_000923_030 |
0.1039143842 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 15 |
Hb_000805_080 |
0.1048978373 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000841_040 |
0.1064202173 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_003053_080 |
0.1066741107 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000039_210 |
0.1071663511 |
- |
- |
PREDICTED: uncharacterized protein LOC105649449 [Jatropha curcas] |
| 19 |
Hb_011068_010 |
0.1080966652 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 20 |
Hb_000029_390 |
0.1084500896 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |