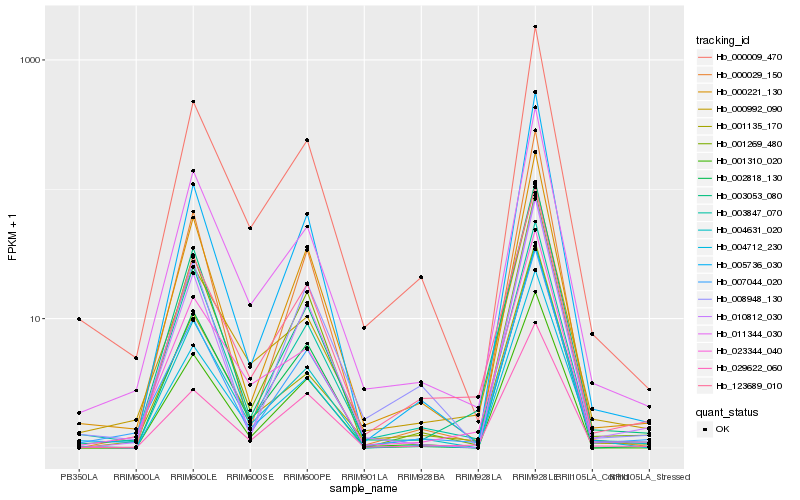
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007044_020 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000221_130 |
0.0584260756 |
- |
- |
thylakoid lumenal 29 kDa protein, chloroplastic [Jatropha curcas] |
| 3 |
Hb_011344_030 |
0.0727123959 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003053_080 |
0.0750366017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010812_030 |
0.0757719838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000009_470 |
0.0778097159 |
- |
- |
Oxygen-evolving enhancer protein 1, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_001269_480 |
0.0798757493 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_029622_060 |
0.0879400799 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004631_020 |
0.0909199193 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 10 |
Hb_001310_020 |
0.0915358916 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_123689_010 |
0.0924775276 |
- |
- |
PREDICTED: uncharacterized protein At1g18480 [Jatropha curcas] |
| 12 |
Hb_005736_030 |
0.0929479472 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 13 |
Hb_002818_130 |
0.0933319019 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_001135_170 |
0.0935461804 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 15 |
Hb_000029_150 |
0.0937866544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000992_090 |
0.0944962782 |
transcription factor |
TF Family: Orphans |
CIL, putative [Ricinus communis] |
| 17 |
Hb_003847_070 |
0.0955440517 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_008948_130 |
0.0968681296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_023344_040 |
0.0975131292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004712_230 |
0.0985420115 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |